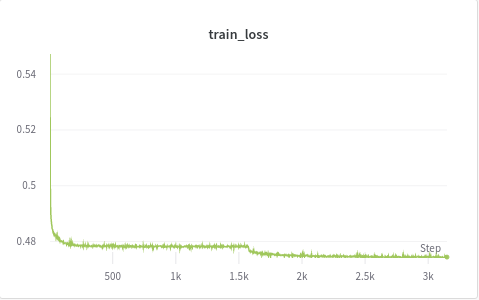
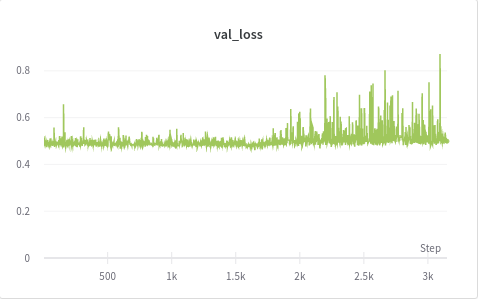
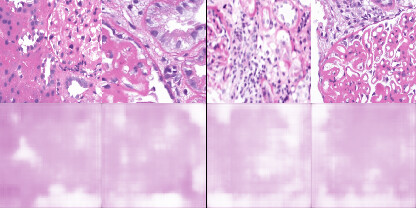
I am new to training autoencoders and trying to work with an unlabeled medical histopathology whole slide image (WSI) dataset and want to visualize possible clusters. The slide image contains a normal tissue region, background, and an abnormality region that I am interested in. I extract patches of fixed size(256x256) from the WSI (2400x1600) and train a convolutional autoencoder. After training, I extract hidden layer (of dimension 32) and perform t-SNE on these feature vectors. But, I am not getting any distinct clusters. A weird thing that I noticed while training is that my binary cross-entropy loss converges within just a few epochs but the reconstructions are really bad. I don’t understand why is that happening. To verify my training, I also tested my model on MNIST and it forms good clusters. Would appreciate any thoughts on this plus any other suggestions on how good is this approach. Below are the training loss, validation loss and reconstructed images.