Hi all,
I use a character-level LSTM model to do binary classification using sequences as input.
The performance is quite good (both f1 score and AUC are around 99%) but I am puzzled by the output probabilities of the model. These probabilities (both in validation and test set) are either extremely close to zero or extremely close to 1 (even equal to one in many cases), meaning the model is extremely confident about its class predictions. To give you a taste of the output probability distribution (similar on both validation and test set):
25% percentile = 0.000054
75% percentile = 0.999986
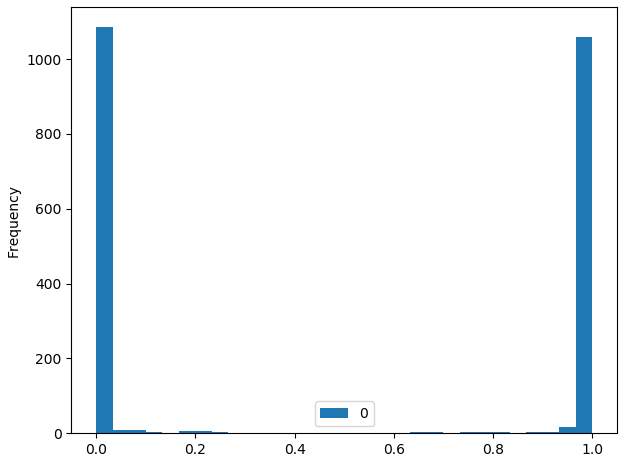
This seems almost too good to be true - the model seems way too confident in almost all cases. I would have expected a much ‘smoother’ distribution of predicted probabilities. It also makes it tricky to set a probability threshold to control for the amount of false positives (since many examples get a predicted probability of 1).
The model is a straightforward implementation of LSTM for classification:
def forward(self, X):
embeds = self.char_embeddings(X)
lstm_out, self.hidden = self.lstm1(embeds, self.hidden)
lstm_out = self.dropout1(lstm_out[:, -1, :])
output = self.dense1(lstm_out)
output = self.sig(output)
return output
Has any one of you experienced something similar before when it comes to binary classification?