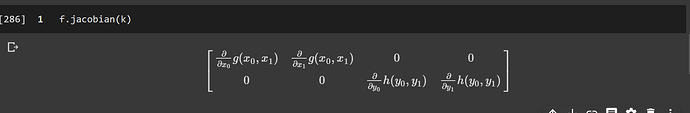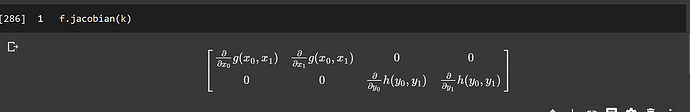when I apply Jacobian in PyTorch autograd (torch==1.5.0), it gives me three dimensional output.
def exp_reducer(x):
return (x**2).sum(dim=1)
inputs = torch.rand(2, 2)
torch.autograd.functional.jacobian(exp_reducer, inputs)
tensor([[[0.3296, 0.9791],
[0.0000, 0.0000]],
[[0.0000, 0.0000],
[1.6706, 0.7365]]])
and
torch.autograd.functional.jacobian(exp_reducer, inputs).shape
(2,2,2)
but I get a 2 dimensional output when using sympy,
x0, y0, x1, y1= sympy.symbols('x0, y0, x1, y1')
f = Matrix([Function('g')(x0, x1), Function('h')(y0, y1)])
k = Matrix([[x0, x1, y0, y1]])
f.jacobian(k)
then it give me, this,
should the shape after applying Jacobian be 2d or 3d?
albanD
April 24, 2020, 5:01pm
2
Hi,
Yes we follow a slightly different semantic.
What sympy is doing here is to linearize the inputs from a (2, 2) Tensor into a (4,) Tensor. And so the Jacobian is (2, 4).
Because we usually allow multiple Tensors, people very often multiple dimensions and linearizing the Tensors can be confusing if you don’t know how they are layed out in memory, we made a different choice here (similar to what tensorflow or jax are doing).
Would it be a good idea to add the symbolic representation to docs?
def pow_adder_reducer(x, y):
print('x: ', x, 'y: ', y)
return (2 * x.pow(2) + 3 * y.pow(2)).sum()
inputs = (torch.rand(2), torch.rand(2))
torch.autograd.functional.hessian(pow_adder_reducer, inputs)
PyTorch gives,
x: tensor([0.4406, 0.9028], grad_fn=<ViewBackward>) y: tensor([0.8790, 0.7052], grad_fn=<ViewBackward>)
((tensor([[4., 0.],
[0., 4.]]), tensor([[0., 0.],
[0., 0.]])), (tensor([[0., 0.],
[0., 0.]]), tensor([[6., 0.],
[0., 6.]])))
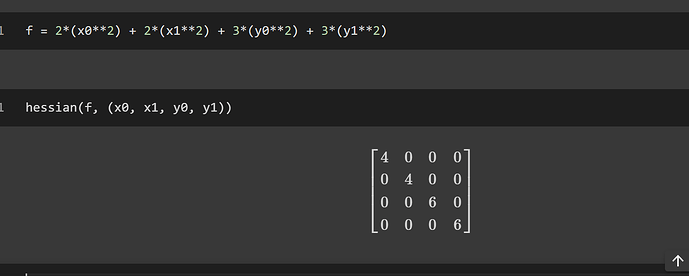
from sympy I get,
is it doing something like,
f1 = 2*(x0**2) + 2*(x1**2)
f2 = 3*(y0**3) + 3*(y1**3)
hessian(f1, (x0, x1))
hessian(f1, (y0, y1))
hessian(f2, (x0, x1))
hessian(f2, (y0, y1))
that results in four 2x2 matrices.
albanD
April 24, 2020, 8:42pm
4
Yes, this is a similar argument. The indexing is interleaved as output tensor, input tensor, output dim, input dim.Add high level autograd functions · Issue #30632 · pytorch/pytorch · GitHub
Would it be a good idea to add the symbolic representation to docs?
Definitely open to suggestions to improve the doc !
Like this one,
and this one,
like this symbolic representation added to docs, this makes it a bit easier to understand when we input >1d tensor.
albanD
April 27, 2020, 6:24pm
6
Thanks, I added it to the issue to work on doc improvements here: https://github.com/pytorch/pytorch/issues/34998