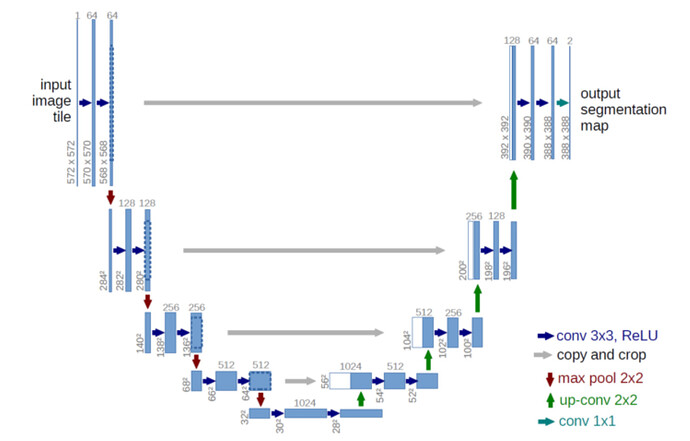
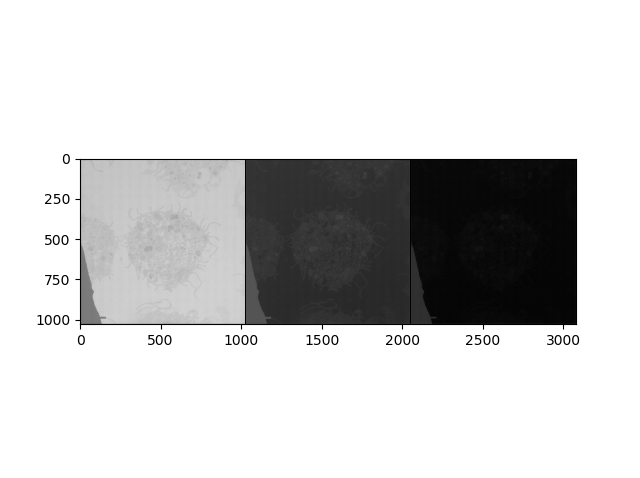
I have one more question. This is the output of your example for visualization in this dataset. Does each of these shows the probability map for each class (num_class = 3)?
Why softmax should apply again while in the last layer of the model already F.log_softmax(self.out(x_up), 1) applied?
In case you are using a log_softmax in your model, you could apply torch.exp to get the probabilities.
Yes, each image should show the probability map for the current class.
hello, I’ve tried your code and I use log_softmax in my model, here is the result I get:
The inputs is:
tensor([[[[ 40., 38., 35., ..., 56., 60., 69.],
[ 27., 30., 33., ..., 69., 60., 65.],
[ 32., 35., 39., ..., 64., 74., 64.],
...,
[ 31., 41., 40., ..., 28., 45., 43.],
[ 36., 40., 44., ..., 32., 36., 50.],
[ 35., 34., 51., ..., 50., 55., 49.]],
[[ 40., 38., 35., ..., 56., 60., 69.],
[ 27., 30., 33., ..., 69., 60., 65.],
[ 32., 35., 39., ..., 64., 74., 64.],
...,
[ 31., 41., 40., ..., 28., 45., 43.],
[ 36., 40., 44., ..., 32., 36., 50.],
[ 35., 34., 51., ..., 50., 55., 49.]],
[[ 40., 38., 35., ..., 56., 60., 69.],
[ 27., 30., 33., ..., 69., 60., 65.],
[ 32., 35., 39., ..., 64., 74., 64.],
...,
[ 31., 41., 40., ..., 28., 45., 43.],
[ 36., 40., 44., ..., 32., 36., 50.],
[ 35., 34., 51., ..., 50., 55., 49.]]]])
The groundtruth label is:
tensor([[[ 0., 0., 0., ..., 1., 1., 1.],
[ 0., 0., 0., ..., 1., 1., 1.],
[ 0., 0., 0., ..., 1., 1., 1.],
...,
[ 0., 0., 0., ..., 1., 1., 1.],
[ 0., 0., 0., ..., 1., 1., 1.],
[ 0., 0., 0., ..., 1., 1., 1.]]])
The output is:
tensor([[[[-0.7782, -0.7635, -0.7635, ..., -0.7635, -0.7635, -0.7740],
[-0.7640, -0.7455, -0.7455, ..., -0.7455, -0.7455, -0.7631],
[-0.7640, -0.7455, -0.7455, ..., -0.7455, -0.7455, -0.7631],
...,
[-0.7640, -0.7455, -0.7455, ..., -0.7455, -0.7455, -0.7631],
[-0.7640, -0.7455, -0.7455, ..., -0.7455, -0.7455, -0.7631],
[-0.7737, -0.7623, -0.7623, ..., -0.7623, -0.7623, -0.7766]],
[[-1.1314, -1.1540, -1.1540, ..., -1.1540, -1.1540, -1.1317],
[-1.1553, -1.1900, -1.1900, ..., -1.1900, -1.1900, -1.1569],
[-1.1553, -1.1900, -1.1900, ..., -1.1900, -1.1900, -1.1569],
...,
[-1.1553, -1.1900, -1.1900, ..., -1.1900, -1.1900, -1.1569],
[-1.1553, -1.1900, -1.1900, ..., -1.1900, -1.1900, -1.1569],
[-1.1364, -1.1588, -1.1588, ..., -1.1588, -1.1588, -1.1345]],
[[-1.5224, -1.5207, -1.5207, ..., -1.5207, -1.5207, -1.5308],
[-1.5175, -1.5083, -1.5083, ..., -1.5083, -1.5083, -1.5173],
[-1.5175, -1.5083, -1.5083, ..., -1.5083, -1.5083, -1.5173],
...,
[-1.5175, -1.5083, -1.5083, ..., -1.5083, -1.5083, -1.5173],
[-1.5175, -1.5083, -1.5083, ..., -1.5083, -1.5083, -1.5173],
[-1.5246, -1.5162, -1.5162, ..., -1.5162, -1.5162, -1.5211]]]], device='cuda:0')
torch.float32
<built-in method size of Tensor object at 0x7f1f0b0b30d8>
The image size is: torch.Size([1, 3, 256, 256]). The inputs data type is: torch.float32
The groundtruth mask size is :torch.Size([1, 256, 256]). The groundtruth mask type is: torch.float32
The predicted mask size is :torch.Size([1, 3, 256, 256]). The predicted mask type is torch.float32
In this case, what does the output (the predicted mask) of the unet represent? I don’t understand why ther are all negative float numbers.
I used mapping that you mentioned before, so the ground truth label is all 0,1,2. In this case, to do the optimisation, I should use which loss function and optimisation?
Thank you so much for your all explanations, that helps me a lot!
The output values are the log probabilities for the current batch.
I.e. each channel in your output represents the log probability of the corresponding class.
If you apply torch.exp on the output, you’ll get the softmax probabilities for each pixel, such that the sum for each pixel location will be 1 (=100%). Since log(1) = 0 and log(0) = -inf you’ll get mostly negative values.
Here is a small dummy example:
x = torch.randn(1, 3, 24, 24)
output = F.log_softmax(x, 1)
print('LogProbs: ', output)
print('Probs: ', torch.exp(output))
print('Prediction: ', torch.argmax(output, 1))
If you are applying F.log_softmax on the model output, you should use nn.NLLLoss as your criterion for a classification/segmentation use case.
I was doing _, predicted = torch.max(outputs.data, 1) to return the indices of max values and get prediction. Would do the same as torch.argmax(output, 1)?
Yes, both calls will return the indices of the max values.
The difference is that torch.max will return both the max values and their indices.
As you can see in your code snippet you are dropping the values and just use the indices.
@ptrblck I am wondering if I add the bottleneck layer to your UNet implementation would be same as this snippet?
basically down4 and center added to your implementation. I just wanted to check with you to make sure I am in right track.
class UNet(nn.Module):
def __init__(self, in_channels, out_channels, n_class, kernel_size, padding, stride):
super(UNet, self).__init__()
self.init_conv = BaseConv(in_channels, out_channels, kernel_size, padding, stride)
self.down1 = DownConv(out_channels, 2 * out_channels, kernel_size, padding, stride)
self.down2 = DownConv(2 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.down3 = DownConv(4 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.down4 = DownConv(8 * out_channels, 16 * out_channels, kernel_size, padding, stride)
self.center = BaseConv( 16 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.up3 = UpConv(8 * out_channels, 4 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.up2 = UpConv(4 * out_channels, 2 * out_channels, 2 * out_channels, kernel_size, padding, stride)
self.up1 = UpConv(2 * out_channels, out_channels, out_channels, kernel_size, padding, stride)
self.out = nn.Conv2d(out_channels, n_class, kernel_size, padding, stride)
def forward(self, x):
# Encoder
x = self.init_conv(x)
x1 = self.down1(x)
x2 = self.down2(x1)
x3 = self.down3(x2)
x4 = self.down4(x3)
x_center = self.center(x4)
# Decoder
x_up1 = self.up3(x_center, x2)
x_up2 = self.up2(x_up1, x1)
x_up3 = self.up1(x_up2, x)
x_out = F.log_softmax(self.out(x_up3), 1)
#print(x_out.size())
return x_out
Yeah, it looks good.
Is the code working as expected?
@ptrblck I’m getting an error. Not sure where I am doing wrong.
x = torch.cat((x, x_skip), dim=1) RuntimeError: invalid argument 0: Sizes of tensors must match except in dimension 1. Got 60 and 30 in dimension 2 at c:\programdata\miniconda3\conda-bld\pytorch_1533100020551\work\aten\src\thc\generic/THCTensorMath.cu:87
@ptrblck I realised I was doing a mistake. If I want to follow the Unet model with your implementation I need to add 4 Encoder , one bridge (bottlenck) and 4 Decoder while I had 4 Encoder and 3 Decoder.
This is the new snippet which is working fine so far but it’s slower than previous one which I assume it’s because of bottleneck. Could you please have a look and let me know am I doing correctly based on the attached UNet model and your implementation.
class UNet(nn.Module):
def __init__(self, in_channels, out_channels, n_class, kernel_size, padding, stride):
super(UNet, self).__init__()
self.init_conv = BaseConv(in_channels, out_channels, kernel_size, padding, stride)
self.down1 = DownConv(out_channels, 2 * out_channels, kernel_size, padding, stride)
self.down2 = DownConv(2 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.down3 = DownConv(4 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.down4 = DownConv(8 * out_channels, 16 * out_channels, kernel_size, padding, stride)
self.center = BaseConv( 16 * out_channels, 16 * out_channels, kernel_size, padding, stride)
self.up4 = UpConv(16 * out_channels, 8 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.up3 = UpConv(8 * out_channels, 4 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.up2 = UpConv(4 * out_channels, 2 * out_channels, 2 * out_channels, kernel_size, padding, stride)
self.up1 = UpConv(2 * out_channels, out_channels, out_channels, kernel_size, padding, stride)
self.out = nn.Conv2d(out_channels, n_class, kernel_size, padding, stride)
def forward(self, x):
# Encoder
x = self.init_conv(x)
x1 = self.down1(x)
x2 = self.down2(x1)
x3 = self.down3(x2)
x4 = self.down4(x3)
x_center = self.center(x4)
# Decoder
x_up1 = self.up4(x_center, x3)
x_up2 = self.up3(x_up1, x2)
x_up3 = self.up2(x_up2, x1)
x_up4 = self.up1(x_up3, x)
x_out = F.log_softmax(self.out(x_up4), 1)
#print(x_out.size())
return x_out
model = UNet(in_channels=1,
out_channels=60,
n_class=2,
kernel_size=3,
padding=1,
stride=1)
model = model.to(device)
@ptrblck I am wondering where should I add dropout layer to the UNet model. It seems some people added dropout after Relu or Conv.
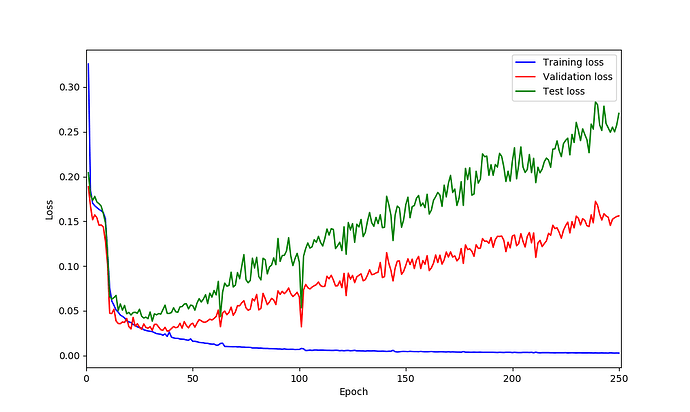
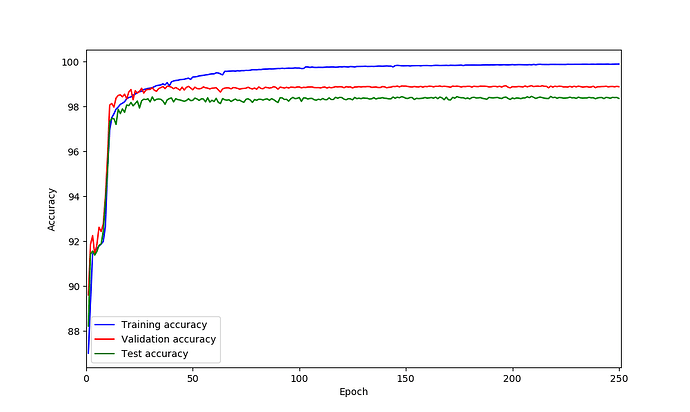
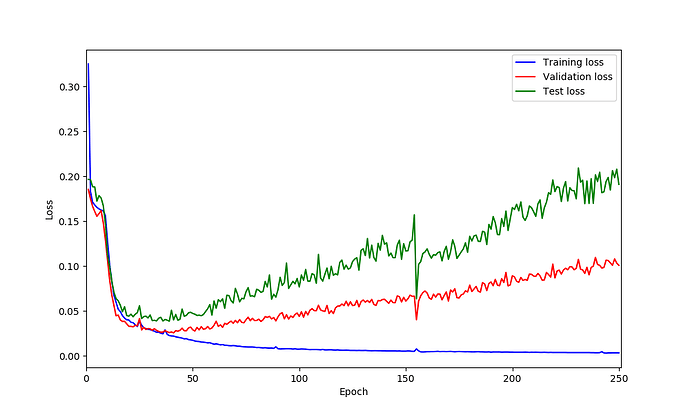
UNet model on my data seems is suffering from over-fitting based on the loss curves. So I was thinking perhaps adding dropout could helps but not sure after which layer should be add? what do you think?
based on your implementation I did added like this but not sure is correct or not.
class BaseConv(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, padding, stride):
super(BaseConv, self).__init__()
self.act = nn.ReLU()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, padding, stride)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, padding, stride)
self.d1 = nn.Dropout(0.2)
def forward(self, x):
x = self.act(self.conv1(x))
x = self.act(self.conv2(x))
x = self.d1(x)
return x
You are right, based on your loss curves it looks like your model is overfitting.
Dropout after the conv layers might be a good idea.
However, I would try to add dropout a bit “later” in the model, i.e. closer to the output, instead of in every BaseConv module. You could add an argument dropout=0.0, which you set higher for later layers.
Let me know, if that helps!
@ptrblck dropout added to the BaseConv module couldn’t help and still loss plots is same as previous experiment which was without dropout.
You said after conv layers and also closer to the output, instead of every BaseConv module. Then, would be fine if add dropout before Relu and immediately after conv?
Yes, this should be alright.
It doesn’t really matter whether dropout is applied before or after ReLU as @rasbt explains here. 
@ptrblck thank you for sharing the link. Now it’s running with dropout. I will post here if it is over-fitting again.
@ptrblck it seems adding dropout could help based on loss curves however I am not sure why do I have those spikes in validation loss! could be because of lr? what do you think?
this is a snippet of network which includes dropout layer:
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.parameters(), lr=0.00001) # 1e-3
class BaseConv(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, padding, stride, droup_rate = 0.2):
super(BaseConv, self).__init__()
self.act = nn.ReLU()
self.conv1 = nn.Conv2d(in_channels, out_channels, kernel_size, padding, stride)
self.d1 = nn.Dropout(droup_rate)
self.conv2 = nn.Conv2d(out_channels, out_channels, kernel_size, padding, stride)
self.d2 = nn.Dropout(droup_rate)
def forward(self, x):
x = self.act(self.conv1(x))
x = self.d1(x)
x = self.act(self.conv2(x))
x = self.d2(x)
return x
class DownConv(nn.Module):
def __init__(self, in_channels, out_channels, kernel_size, padding, stride):
super(DownConv, self).__init__()
self.pool1 = nn.MaxPool2d(kernel_size=2)
self.conv_block = BaseConv(in_channels, out_channels, kernel_size, padding, stride)
def forward(self, x):
x = self.pool1(x)
x = self.conv_block(x)
return x
class UpConv(nn.Module):
def __init__ (self, in_channels, in_channels_skip, out_channels, kernel_size, padding, stride):
super(UpConv, self).__init__()
self.conv_trans1 = nn.ConvTranspose2d(in_channels, in_channels, kernel_size=2, padding=0, stride=2)
self.conv_block = BaseConv(in_channels=in_channels + in_channels_skip, out_channels= out_channels, kernel_size=kernel_size, padding=padding, stride=stride)
def forward(self, x, x_skip):
x = self.conv_trans1(x)
x = torch.cat((x, x_skip), dim=1)
x= self.conv_block(x)
return x
class UNet(nn.Module):
def __init__(self, in_channels, out_channels, n_class, kernel_size, padding, stride, droup_rate = 0.2):
super(UNet, self).__init__()
self.init_conv = BaseConv(in_channels, out_channels, kernel_size, padding, stride)
self.d3 = nn.Dropout(droup_rate)
self.down1 = DownConv(out_channels, 2 * out_channels, kernel_size, padding, stride)
self.down2 = DownConv(2 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.down3 = DownConv(4 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.down4 = DownConv(8 * out_channels, 16 * out_channels, kernel_size, padding, stride)
self.center = BaseConv( 16 * out_channels, 16 * out_channels, kernel_size, padding, stride)
self.d4 = nn.Dropout(droup_rate)
self.up4 = UpConv(16 * out_channels, 8 * out_channels, 8 * out_channels, kernel_size, padding, stride)
self.up3 = UpConv(8 * out_channels, 4 * out_channels, 4 * out_channels, kernel_size, padding, stride)
self.up2 = UpConv(4 * out_channels, 2 * out_channels, 2 * out_channels, kernel_size, padding, stride)
self.up1 = UpConv(2 * out_channels, out_channels, out_channels, kernel_size, padding, stride)
self.d5 = nn.Dropout(droup_rate)
self.out = nn.Conv2d(out_channels, n_class, kernel_size, padding, stride)
def forward(self, x):
# Encoder
x = self.init_conv(x)
d3 = self.d3(x)
x1 = self.down1(d3)
x2 = self.down2(x1)
x3 = self.down3(x2)
x4 = self.down4(x3)
x_center = self.center(x4)
d4 = self.d4(x_center)
# Decoder
x_up1 = self.up4(d4, x3)
x_up2 = self.up3(x_up1, x2)
x_up3 = self.up2(x_up2, x1)
x_up4 = self.up1(x_up3, x)
d5 = self.d5(x_up4)
x_out = F.log_softmax(self.out(d5), 1)
#print(x_out.size())
return x_out
model = UNet(in_channels=1,
out_channels=64,
n_class=2,
kernel_size=3,
padding=1,
stride=1)
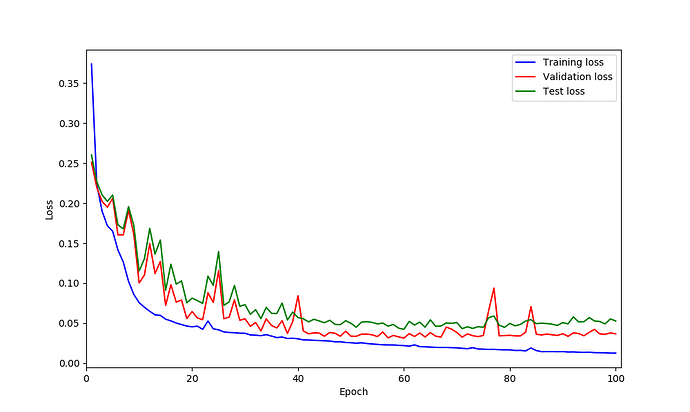
this is loss curves:
I’m not sure where these spikes come from and they don’t look too bad in my opinion.
Are you changing the learning rate using a scheduler?
@ptrblck No, at the moment is like optimizer = optim.Adam(model.parameters(), lr=0.00001) which is fix lr. I’ll try to do scheduler as well.
self.mapping = {
85: 0,
170: 1,
255: 2
}
In this what are 85,170 and 255 ? they are color codes of mask image ?
If i have black background and green and blue colors in my mask image. Then how can i define mapping ?
Can we use a hex value for the mapping?
example: self.mapping = {
(85, 64, 64): 0,
(45, 0, 170): 1,
(255, 255, 86): 2
}