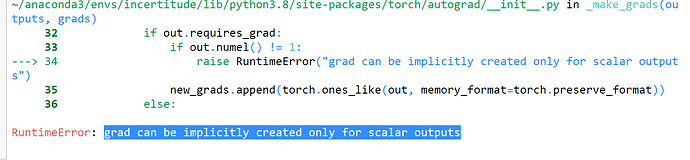
I am building a MLP with 2 outputs as mean and variance because, I am working on quantifying uncertainty of the model. I have used a proper scoring for NLL for regression as metrics. My training function passed with MSE loss function but when I am applying my proper scoring function , I am getting the following error :
Here is a piece of my function:
def train1(model, epoch, trainloader, criterion, sp, optimizer):
model.train()
for batch_idx, (data, target) in enumerate(trainloader):
if torch.cuda.is_available():
data, target = data.cuda(), target.cuda()
optimizer.zero_grad()
output = model(data)
print(output)
mu, sig = output[0][0], sp(output[0][1])+(10)**-6
loss = nll_criterion(mu, sig, target)
loss = criterion(output,target)
loss.backward()
optimizer.step()
One more thing. My code passed when I do not use dataloader or any pytorch method, like tensor.
But when I am using Dataloader and convert my variables to tensors. I am having troubles - I would like to use pytorch for this project.
def main():
n
n_hidden = 100
batch_size = 20
print("Setting up data")
dict_ = split_data(df,data_ratio=0.30)
X_train, Y_train, = dict_["train_x"].values.astype(np.float32), dict_["train_y"].values.astype(np.float32)
X_test, Y_test, = dict_["test_x"].values.astype(np.float32), dict_["test_y"].values.astype(np.float32)
#dataloader
trainset = mydata(X_train, Y_train)
trainloader = DataLoader(trainset,shuffle=False,batch_size=20)
testset = mydata(X_test, Y_test)
testloader = DataLoader(testset,shuffle=False,batch_size=20)
trainset = torch.utils.data.TensorDataset(torch.from_numpy(X_train),torch.from_numpy(Y_train))
trainloader = torch.utils.data.DataLoader(trainset, batch_size=20, shuffle=True, num_workers=2)
testset = torch.utils.data.TensorDataset(torch.from_numpy(X_test),torch.from_numpy(Y_test))
testloader = torch.utils.data.DataLoader(testset, batch_size=1, shuffle=False, num_workers=2)
model = make_model(X_train, n_hidden)
optimizer = optim.Adam(model.parameters(), lr=0.001)
criterion = nn.MSELoss()
# Proper scoring rule using negative log likelihood scoring rule
nll_criterion = lambda mu, sigma, y: torch.log(sigma)/2 + ((y-mu)**2)/(2*sigma)
sp = torch.nn.Softplus()
n_epochs= 1000
predict_every = 15
running_loss = []
for epoch in range(n_epochs):
epoch_loss = 0
print("(Start) Epoch ",epoch," of ",n_epochs,":")
epoch_loss = train1(model, epoch, trainloader, criterion=nll_criterion,sp=sp, optimizer=optimiz