Hi @erroneus,
Finally got some time to dig through your DIM code and the paper. Very impressive and very thorough.
The code is obviously highly modular, but it makes it a little difficult to track how the DIM network gets constructed and the loss calculated. I made a small, hard-coded network to implement the local/global DIM loss calculation for a specific tensor shape. This was written from your posted DIM code and the various figures in the paper (and appendix). Can you take a look and let me know if I’m understanding things correctly?
(I’m fairly new to all this, so apologies for any obviously dumb mistakes)
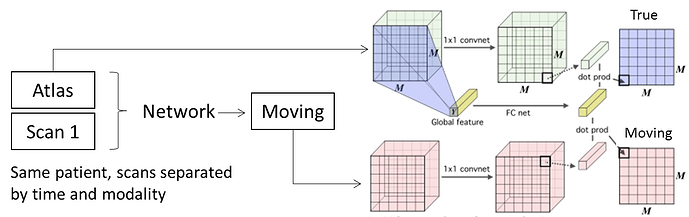
Input A & B: 256 x 256 x 256 tensor representing a 3D image volume (“Atlas” and “Moving” volumes)
Volumes are greyscale (single channel).
When I use actual image data, the network trains, and seems to generate numbers that accurately correlate with hard-calculated mutual information. Which is fantastic, but only if I’m actually doing this right. Appreciate any insight/critiques/etc. Thanks!
A = torch.randint(0, 256, (2, 1, 256, 256, 256)).float().requires_grad_(True)
B = torch.randint(0, 256, (2, 1, 256, 256, 256)).float().requires_grad_(True)
def get_positive_expectation(p_samples, average=True):
#Measure = JSD (Simplified from DIM Code for clarity)
log_2 = math.log(2.)
Ep = log_2 - F.softplus(-p_samples) # Note JSD will be shifted
if average:
return Ep.mean()
else:
return Ep
def get_negative_expectation(q_samples, average=True):
#Measure = JSD (Simplified from DIM Code for clarity)
log_2 = math.log(2.)
Eq = F.softplus(-q_samples) + q_samples - log_2 # Note JSD will be shifted
if average:
return Eq.mean()
else:
return Eq
def loss_calc(lmap, gmap):
#The fenchel_dual_loss from the DIM code
#Reshape tensors dims to (N, Channels, chunks)
lmap = lmap.reshape(2,128,-1)
gmap = gmap.squeeze()
N, units, n_locals = lmap.size()
n_multis = gmap.size(2)
# First we make the input tensors the right shape.
l = lmap.view(N, units, n_locals)
l = lmap.permute(0, 2, 1)
l = lmap.reshape(-1, units)
m = gmap.view(N, units, n_multis)
m = gmap.permute(0, 2, 1)
m = gmap.reshape(-1, units)
u = torch.mm(m, l.t())
u = u.reshape(N, n_multis, N, n_locals).permute(0, 2, 3, 1)
mask = torch.eye(N).to(l.device)
n_mask = 1 - mask
E_pos = get_positive_expectation(u, average=False).mean(2).mean(2)
E_neg = get_negative_expectation(u, average=False).mean(2).mean(2)
E_pos = (E_pos * mask).sum() / mask.sum()
E_neg = (E_neg * n_mask).sum() / n_mask.sum()
loss = E_neg - E_pos
return loss
class Mixed_Dim(torch.nn.Module):
def __init__(self):
super(Mixed_Dim, self).__init__()
#Local Feature Map: Local feature map of size [N, 128, 8, 8, 8]
self.lmapnet = Sequential(Conv3d(1, 8, 2, 2), ReLU(), BatchNorm3d(8),
Conv3d(8, 16, 2, 2), ReLU(), BatchNorm3d(16),
Conv3d(16, 32, 2, 2), ReLU(), BatchNorm3d(32),
Conv3d(32, 64, 2, 2), ReLU(), BatchNorm3d(64),
Conv3d(64, 128, 2, 2), ReLU(), BatchNorm3d(128)
)
#Global Feature Map: Global feature map of size [N, 128, 1, 1, 1]
self.gmapnet = Sequential(Conv3d(128, 128, 2, 2), ReLU(), BatchNorm3d(128),
Conv3d(128, 128, 2, 2), ReLU(), BatchNorm3d(128),
Conv3d(128, 128, 2, 2), ReLU()
)
#Per paper, global map is activated:
self.gfc1 = Sequential(Linear(1, 512), ReLU(), BatchNorm3d(128),
Linear(512, 512))
self.gfc2 = Sequential(Linear(1, 512), ReLU(), BatchNorm3d(128))
#Per paper, local map is activated:
self.lfc1 = Sequential(Conv3d(128, 128, 1, 1), ReLU(), BatchNorm3d(128),
Conv3d(128, 128, 1, 1))
self.lfc2 = Sequential(Conv3d(128, 128, 1, 1), ReLU(), BatchNorm3d(128))
self.Laynorm = LayerNorm([128, 8, 8, 8])
def forward(self, moving, atlas):
#Local feature maps of moving and atlas
lmap_moving = self.lmapnet(moving)
lmap_atlas = self.lmapnet(atlas)
#Global feature map of atlas
gmap_atlas = self.gmapnet(lmap_atlas)
#Encode Global feature map
gout1, gout2 = self.gfc1(gmap_atlas), self.gfc2(gmap_atlas)
gmap_atlas_enc = self.gfc1(gmap_atlas) + self.gfc2(gmap_atlas)
#Encode Local feature maps
lmap_atlas_enc = self.Laynorm(self.lfc1(lmap_atlas) + self.lfc2(lmap_atlas))
lmap_moving_enc = self.Laynorm(self.lfc1(lmap_moving) + self.lfc2(lmap_moving))
return lmap_atlas_enc, lmap_moving_enc, gmap_atlas_enc
model_dim = Mixed_Dim().cuda().train()
optim = torch.optim.Adam(model_dim.parameters(), lr=0.01)
A = A.cuda()
B = B.cuda()
for epoch in range(100):
loc_atlas, loc_moving, glob_atlas = model_dim(B, A)
loss1 = loss_calc(loc_moving, glob_atlas)
loss2 = loss_calc(loc_atlas, glob_atlas)
loss = (loss2 + loss1)
model_dim.zero_grad()
print(loss.item())
loss.backward()
optim.step()