Hi:
I am trying to implement an generative poisoning attack method and the paper can be found here: [1703.01340] Generative Poisoning Attack Method Against Neural Networks. So the problem I am having is very tricky and let me explain it step by step:
Repeat:
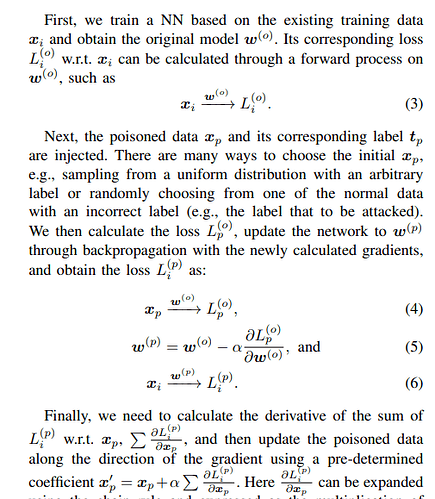
Step 1: I trained a resnet18 with animal images from 4 classes.
Step 2: I feed a random image(I call it Xp) into the network(This is the poisoned image init and I am updating) and get a loss_p.
Step 3: Update model parameter based on loss_p to get w_p(The updated parameter after feeding in Xp)
Step 4: Feed the original training data into W_p and get a loss_p_i.(Note that the goal is to maximize this loss_p_i)
Step 5: Update Xp by adding lr*dloss_p_i/dxp
Here is a clip of the paper I am following:
My problem is that, I am updating dloss_p_i/dxp. However dloss_p_i is not directly related. dloss_p_i is related to W_p and W_p is related to Xp. Therefore when I calculate dloss_p_i/dxp I get error messages like this:
One of the differentiated Tensors appears to not have been used in the graph. Set allow_unused=True if this is the desired behavior.
Here is the code:
optimizer_ft = optim.SGD(resnet.parameters(), lr=0.001, momentum=0.9)
criterion = nn.CrossEntropyLoss()
max_poison_iter = 10
# Change the label to an inccorect one
if poison_label[0] == 0:
poison_label[0] = 1
else:
poison_label[0] = 0
for i in range(max_poison_iter):
print(i)
for images, labels in trainset_loader:
images = images.to(device)
labels = labels.to(device)
resnet = resnet.to(device)
p_outputs = resnet(poison_image)
p_loss = criterion(p_outputs, poison_label)
p_loss.backward(create_graph = True, retain_graph = True)
optimizer_ft.step()
o_outputs = resnet(images)
loss_p_i = criterion(o_outputs, labels)
p_grad = grad(loss_p_i, poison_image)
poison_image.data += 0.1*p_grad[0].sign()