Hi @Surbhi_Khushu,
You can achieve that using TorchIO:
from pathlib import Path
import torchio as tio
out_dir = Path('/tmp/patches')
out_dir.mkdir(exist_ok=True)
patch_size = 64
patch_overlap = 18
subject = tio.Subject(
ct=tio.Image('/tmp/ct.nii.gz', type=tio.INTENSITY),
mr=tio.Image('/tmp/mr.nii.gz', type=tio.INTENSITY),
)
sampler = tio.data.GridSampler(
subject,
patch_size,
patch_overlap,
)
for i, patch in enumerate(sampler):
patch.ct.save(out_dir / f'ct_{i}.nii.gz')
patch.mr.save(out_dir / f'mr_{i}.nii.gz')
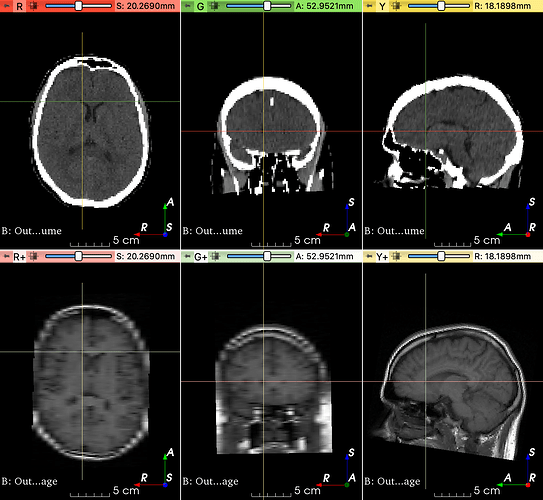
Full volumes:
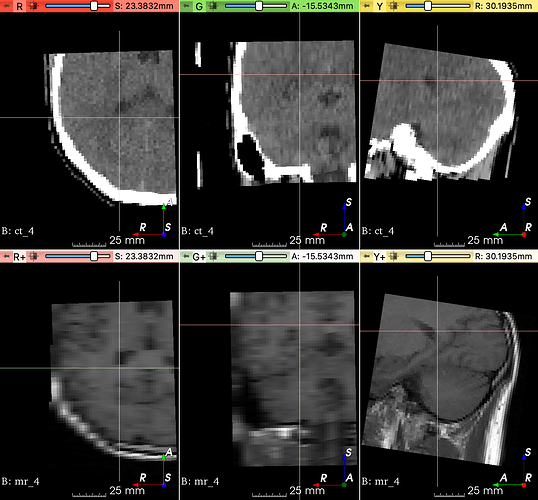
One of the extracted patches:
Take a look at the documentation for patch-based pipelines.