Hello @ptrblck,
Can you please review the following snippet for where I might be going wrong?
model = back_bone_model # A 4 Layered CONV model.
meta_opt = torch.optim.Adam(model.parameters(), lr=1e-3)
loss_fn = nn.CrossEntropyLoss(reduction='mean')
for iteration in range(iterations):
train_loss, train_acc = 0, 0
for task in range(meta_batch_size):
# Copy model parameters. You don't want to train on model in inner loop :)
learner = deepcopy(model)
batch = tasksets.train.sample()
loss_val, acc_val = inner_loop(learner, batch, loss_fn, 1, device) # Gradient steps: 1
train_loss += loss_val/meta_batch_size
train_acc += acc_val/meta_batch_size
del learner
train_loss.backward()
meta_opt.step()
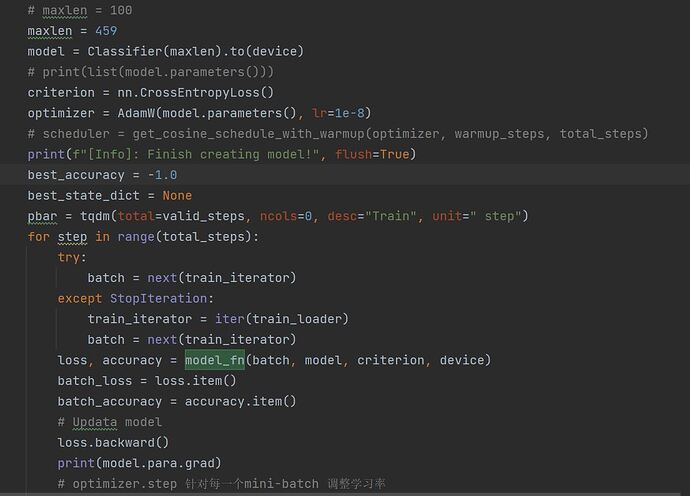
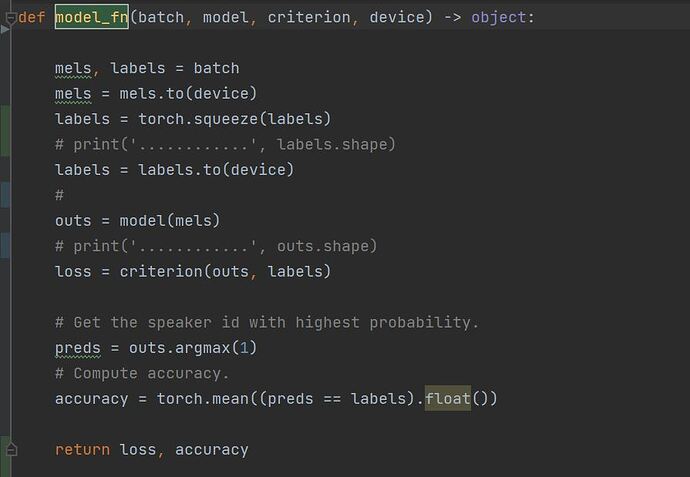
A few details on the inner_loop().
I am taking a few samples (a batch) and updating the parameters of learner() in the inner_loop(). I am accumulating the loss over a few iterations of these batches, and then updating the weights of model() (main model) using the accumulated loss.
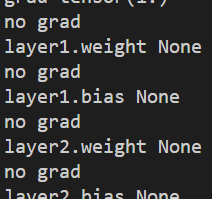
Now, the graph seems to be breaking somewhere in between, as model parameters have .grad as None (for all parameters). (Meaning, that train_loss.backward() is not updating weights. I am not able to figure out where I might be going wrong, thus, I would really appreciate if you are able to point me the mistake in my code. Thank you!