I define my Dataset class:
class MyDataset(Dataset):
def __init__(self, images, n, labels=None, transforms=None):
self.X = images
self.y = labels
self.n = n
self.transforms = transforms
def __len__(self):
return (len(self.X))
def __getitem__(self, i):
data = self.X.iloc[i, :]
# print(data.shape)
data = np.asarray(data).astype(np.float).reshape(1,n,n)
if self.transforms:
data = self.transforms(data).reshape(1,n,n)
if self.y is not None:
y = self.y.iloc[i,:]
# y = np.asarray(y).astype(np.float).reshape(2*n+1,) # for 257-vector of labels
y = np.asarray(y).astype(np.float).reshape(128,) # for 128-vector of labels
return (data, y)
else:
return data
Then I create the instances of the train, dev, and test data:
train_data = MyDataset(train_images, n, train_labels, None)
dev_data = MyDataset(dev_images, n, dev_labels, None)
test_data = MyDataset(test_images, n, test_labels, None)
After having trained my network, I want to see how well it behaves with the test_data. This is what I do to check this.
network = Network(128).double()
btch_sz = 100 # batch size (e.g. 100)
loader_test = DataLoader(test_data_normal,batch_size=btch_sz,shuffle=True,num_workers=0)
for data in loader_test:
images_test, labels_test = data
labels_test = labels_test.detach().numpy()
# to get the data samples from the first batch I break the for-loop cycle
break
outputs_test = network(images_test)
outputs_test = outputs_test.detach().numpy()
Then I plot comparative graphs comparing predicted outputs_test with ground-truth labels_test:
nnn = 3 # to chose a specific sample from the first batch of the test dataset (e.g. 3rd data sample in the batch of 100 samples)
figure, ax1 = plt.subplots()
ax1.plot(labels_test[nnn-1,:], color=u'#1f77b4')
ax1.plot(160*outputs_test[nnn-1,:], color=u'#ff7f0e'')
ax1.legend(['True','Predicted'])
ax1.set_ylim([-40,40])
As you can see, I have to multiply the outputs_test by 160 to make the resulting curve comparable in amplitude with the ground-truth curve. This is the first question, why would it be so?
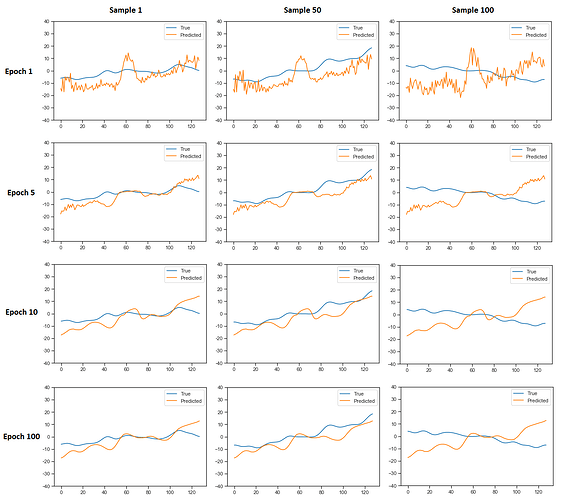
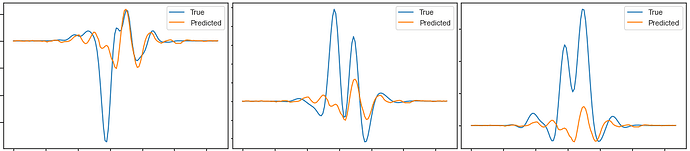
Here are the plots for different data-samples from the batch of 100 and for different epochs of the training (the rows represents indicated epochs and the columns represent indicated data-samples from the batch):
As you can see, the result doesnt depend much on what data-sample is fed into the network. This is another question, why is that so?
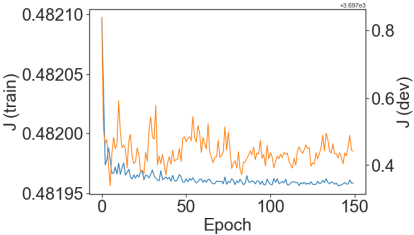
Addon: For the reference this is how cost evolved over 150 epochs.
Also, my network consists of 10 convolutional layers (including batchnorms and maxpools in some places) and 5 linear fully-connected layers.
This is the output of the print(network):
Network(
(encoder): MyEncoder(
(conv_blocks): Sequential(
(0): Sequential(
(0): Conv2d(1, 6, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(1): Sequential(
(0): Conv2d(6, 12, kernel_size=(5, 5), stride=(1, 1))
(1): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(2): BatchNorm2d(12, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(3): ReLU()
)
(2): Sequential(
(0): Conv2d(12, 32, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(3): Sequential(
(0): Conv2d(32, 32, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(4): Sequential(
(0): Conv2d(32, 16, kernel_size=(5, 5), stride=(1, 1))
(1): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(2): BatchNorm2d(16, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(3): ReLU()
)
(5): Sequential(
(0): Conv2d(16, 16, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(6): Sequential(
(0): Conv2d(16, 14, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(7): Sequential(
(0): Conv2d(14, 14, kernel_size=(5, 5), stride=(1, 1))
(1): BatchNorm2d(14, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU()
)
(8): Sequential(
(0): Conv2d(14, 12, kernel_size=(5, 5), stride=(1, 1))
(1): ReLU()
)
(9): Sequential(
(0): Conv2d(12, 12, kernel_size=(5, 5), stride=(1, 1))
(1): MaxPool2d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
(2): BatchNorm2d(12, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(3): ReLU()
)
)
)
(decoder): MyDecoder(
(fc_blocks): Sequential(
(0): Sequential(
(0): Linear(in_features=48, out_features=120, bias=True)
(1): ReLU()
)
(1): Sequential(
(0): Linear(in_features=120, out_features=60, bias=True)
(1): ReLU()
)
(2): Sequential(
(0): Linear(in_features=60, out_features=40, bias=True)
(1): BatchNorm1d(40, eps=1e-05, momentum=0.1, affine=True, track_running_stats=True)
(2): ReLU()
)
(3): Sequential(
(0): Linear(in_features=40, out_features=20, bias=True)
(1): ReLU()
)
(4): Sequential(
(0): Linear(in_features=20, out_features=10, bias=True)
(1): ReLU()
)
)
)
(last): Linear(in_features=10, out_features=128, bias=True)
)