Now that pytorch can autograd, why we still need to define backward when defining customer function
You don’t need to define the backward function, of you use just PyTorch code.
Once you need another library, autograd cannot create the backward pass for you.
got it, thank you very much
but if I comment the definition of backward function, then it sends me the error message " raise NotImplementedError
NotImplementedError"
the following is my code
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(Function):
@staticmethod
def forward(self,input,sigma,center):
dist1=center-input
self.dist1=dist1
dist=torch.pow(dist1,2)
dist2=torch.sum(dist,1)
self.dist2=dist2
sigma2=torch.pow(sigma,2)
self.sigma2=sigma2
dist=-1/2*sigma2*dist2
output=torch.exp(dist)
self.save_for_backward(input,sigma,center,output)
return output
"""
@staticmethod
def backward(self,grad_output):
input,sigma,center,output=self.saved_variables
grad_input=grad_sigma=grad_center=None
if self.needs_input_grad[0]:
tmp=grad_output*output*self.sigma2
grad_input=torch.mv(center.t(),tmp)
if self.needs_input_grad[1]:
grad_sigma=-grad_output*output*sigma*self.dist2
if self.needs_input_grad[2]:
tmp = -grad_output*output*self.sigma2
tmp = torch.diag(tmp)
grad_center = torch.mm(tmp,self.dist1)
return grad_input,grad_sigma,grad_center
"""
from torch.autograd import gradcheck
input =(Variable(torch.randn(10).double(), requires_grad=True), Variable(torch.randn(20).double(), requires_grad=True),
Variable(torch.randn(20,10).double(), requires_grad=True))
test = gradcheck(Gaussian.apply, input, eps=1e-6, atol=1e-4)
print(test)
It looks like some arguments are still from 0.1.12, e.g. the self argument, while others are from the current version, e.g. staticmethod. Could you update your code using this example?
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Tue Apr 10 10:04:34 2018
@author: apple
"""
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(Function):
@staticmethod
def forward(ctx,input,sigma,center):
dist1=center-input
ctx.dist1=dist1
dist=torch.pow(dist1,2)
dist2=torch.sum(dist,1)
ctx.dist2=dist2
sigma2=torch.pow(sigma,2)
ctx.sigma2=sigma2
dist=-1/2*sigma2*dist2
output=torch.exp(dist)
ctx.save_for_backward(input,sigma,center,output)
return output
"""
@staticmethod
def backward(ctx,grad_output):
input,sigma,center,output=ctx.saved_variables
grad_input=grad_sigma=grad_center=None
if ctx.needs_input_grad[0]:
tmp=grad_output*output*ctx.sigma2
grad_input=torch.mv(center.t(),tmp)
if ctx.needs_input_grad[1]:
grad_sigma=-grad_output*output*sigma*ctx.dist2
if ctx.needs_input_grad[2]:
tmp = -grad_output*output*ctx.sigma2
tmp = torch.diag(tmp)
grad_center = torch.mm(tmp,ctx.dist1)
return grad_input,grad_sigma,grad_center
"""
from torch.autograd import gradcheck
input =(Variable(torch.randn(10).double(), requires_grad=True), Variable(torch.randn(20).double(), requires_grad=True),
Variable(torch.randn(20,10).double(), requires_grad=True))
test = gradcheck(Gaussian.apply, input, eps=1e-6, atol=1e-4)
print(test)
it doesn’t work even I changed self to ctx
You are mixing two difference things:
- Writing own module using autograd (this you want)
- Extending autograd by your own function, because ex. torch doesn’t have Bernoulli distribution (you are doing that)
So if you want to create Gaussian distribution and torch have enough function to code it, you should use nn.Module as a base (Function require to have backward pass coded by user)
Also in such case you are able to test gradient, but it would return True because autograd calculate it:)
So, in summary, use nn.Module if you are not using any special-external library (like scipy, nlp etc.)
So code for your function should look like that:
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
from torch.autograd import gradcheck
class Gaussian(nn.Module):
def forward(self, input,sigma,center):
dist1=center-input
dist=torch.pow(dist1,2)
dist2=torch.sum(dist,1)
sigma2=torch.pow(sigma,2)
dist=-1/2*sigma2*dist2
output=torch.exp(dist)
return output
input =(Variable(torch.randn(10).double(), requires_grad=True), Variable(torch.randn(20).double(), requires_grad=True),
Variable(torch.randn(20,10).double(), requires_grad=True))
test = gradcheck(Gaussian().forward, input, eps=1e-6, atol=1e-4)
print(test)
# for to use this module
g = Gaussian()
data = g(*input)
loss = data.sum() # need to be a scalar
loss.backward()
I still got error even I followed your coding, the following is the code and the error message is among it
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Tue Apr 10 10:04:34 2018
@author: apple
"""
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(nn.Module):
def __init__(ctx,in_features,out_features):
super(Gaussian,ctx).__init__()
ctx.sigma=nn.Parameter(torch.Tensor(out_features))
ctx.center=nn.Parameter(torch.Tensor(out_features,in_features))
def forward(ctx,input):
dist=ctx.center-input
"""
sub() received an invalid combination of arguments - got (torch.FloatTensor), but expected one of:
* (float other, float alpha)
* (Variable other, float alpha)
"""
dist=torch.pow(dist,2)
dist=torch.sum(dist,1)
sigma=torch.pow(ctx.sigma,2)
dist=-1/2*sigma*dist
output=torch.exp(dist)
return output
class RBF(nn.Module):
def __init__(ctx,in_features,out_features):
super(RBF,ctx).__init__()
ctx.Gaussian1 = Gaussian(in_features,out_features)
def forward(ctx,x):
return ctx.Gaussian1(x)
def Gaussian_init(m):
if isinstance(m, Gaussian):
m.sigma=nn.Parameter(torch.Tensor([3]))
m.center=nn.Parameter(torch.Tensor([[1,2]]))
model=RBF(2,1)
model.apply(Gaussian_init)
x=torch.Tensor([1,2])
y=model(x)
I want to define a new layer Gaussian, and to use that layer to build a net. And I want to apply the Gaussian_init to initiate the parameters in Gaussian
As @melgor said, you are still mixing Module (collection of autograd operations) and Function (a custom autograd option).
Read these:
Function: http://pytorch.org/tutorials/beginner/pytorch_with_examples.html#pytorch-defining-new-autograd-functions
Module: http://pytorch.org/tutorials/beginner/pytorch_with_examples.html#pytorch-custom-nn-modules
I am new to pytorch, can you do me a favor, I hope you could figure out more specifically, thanks
I followed your code, but still got error, can you do me a favor
First of all, learn some Python first.
Stop using ctx, use self
Second stuff is that in PyTorch every element of dynamic-graph need to be a Variable or Parameter. So input need also be a Variable (if you don’t get it, watch some tutorials about PyTorch)
My working code is here, but it is still not perfect, initialization of Gaussian is horrible (it should be done in different way)
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Tue Apr 10 10:04:34 2018
@author: apple
"""
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(nn.Module):
def __init__(self,in_features,out_features):
super(Gaussian,self).__init__()
self.sigma=nn.Parameter(torch.Tensor(out_features))
self.center=nn.Parameter(torch.Tensor(out_features,in_features))
def forward(self, input):
dist=self.center-input
dist=torch.pow(dist,2)
dist=torch.sum(dist,1)
sigma=torch.pow(self.sigma,2)
dist=-1/2*sigma*dist
output=torch.exp(dist)
return output
class RBF(nn.Module):
def __init__(self,in_features,out_features):
super(RBF,self).__init__()
self.Gaussian1 = Gaussian(in_features,out_features)
def forward(self,x):
return self.Gaussian1(x)
def Gaussian_init(m):
if isinstance(m, Gaussian):
m.sigma=nn.Parameter(torch.Tensor([3]))
m.center=nn.Parameter(torch.Tensor([[1,2]]))
model=RBF(2,1)
model.apply(Gaussian_init)
x=Variable(torch.Tensor([1,2]))
y=model(x)
print(y)
Thanks for your code, and it works.
But the problem occurs again when I want to add a Linear layer onto my net.
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Tue Apr 10 10:04:34 2018
@author: apple
"""
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(nn.Module):
def __init__(self,in_features,out_features):
super(Gaussian,self).__init__()
self.sigma=nn.Parameter(torch.Tensor(out_features))
self.center=nn.Parameter(torch.Tensor(out_features,in_features))
def forward(self,input):
dist=self.center-input
dist=torch.pow(dist,2)
dist=torch.sum(dist,1)
sigma=torch.pow(self.sigma,2)
dist=-1/2*sigma*dist
output=torch.exp(dist)
return output
class RBF(nn.Module):
def __init__(self,in_features,h_features,out_features):
super(RBF,self).__init__()
self.linear = nn.Linear(in_features,h_features,bias=False)
self.Gaussian1 = Gaussian(h_features,out_features)
def forward(self,x):
x= self.linear(x)
x = Gaussian(x)
return x
def Weight_init(m):
if isinstance(m,nn.Linear):
m.weight= nn.Parameter(torch.Tensor([[1,2,3],[4,5,6]]))
def Gaussian_init(m):
if isinstance(m, Gaussian):
m.sigma=nn.Parameter(torch.Tensor([3]))
m.center=nn.Parameter(torch.Tensor([[1,2]]))
model=RBF(3,2,1)
model.apply(Weight_init)
model.apply(Gaussian_init)
x=Variable(torch.Tensor([3,2]))
y=model(x)
print(y)
The error message is:
" size mismatch, m1: [1 x 2], m2: [3 x 2] at /Users/soumith/code/builder/wheel/pytorch-src/torch/lib/TH/generic/THTensorMath.c:1416"
And I searched on http://pytorch.org/tutorials/beginner/pytorch_with_examples.html , and it puzzled me a lot
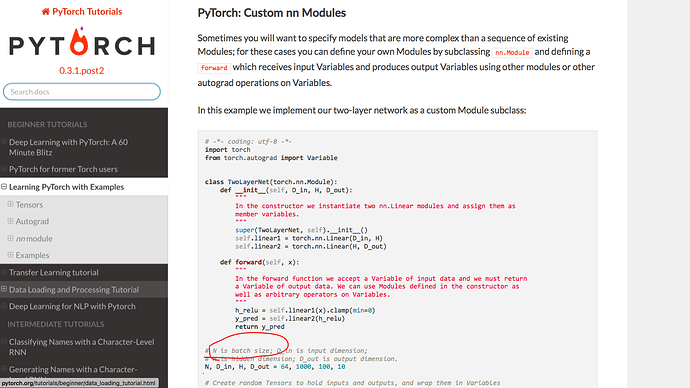
In the Linear, we need to consider the batch size. My code is simple , but I want to use SGD later. Then, do I need to consider the batch_size when I define my own Gaussian layer?
Do I need to consider the batch size?
@king_wang pls trace back the error popping up in your piece of code. size mismatch, m1:[1 x 2], m2: [3 x 2] is very clear that the dimensionality does not match. Note that model = RBF(3,2,1) means the dimensionality of input feature should be 3 while your script torch.Tensor([3,2]) just alloc a tensor [3,2].
Ok, thanks! I thought the problem was because I didn’t consider the batch_size, but it still doesn’t work well
#!/usr/bin/env python3
# -*- coding: utf-8 -*-
"""
Created on Tue Apr 10 10:04:34 2018
@author: apple
"""
import torch
import torch.nn as nn
from torch.autograd import Variable
from torch.autograd import Function
class Gaussian(nn.Module):
def __init__(self,in_features,out_features):
super(Gaussian,self).__init__()
self.sigma=nn.Parameter(torch.Tensor(out_features))
self.center=nn.Parameter(torch.Tensor(out_features,in_features))
def forward(self,input):
dist=self.center-input
dist=torch.pow(dist,2)
dist=torch.sum(dist,1)
sigma=torch.pow(self.sigma,2)
dist=-1/2*sigma*dist
output=torch.exp(dist)
return output
class RBF(nn.Module):
def __init__(self,in_features,h_features,out_features):
super(RBF,self).__init__()
self.linear = nn.Linear(in_features,h_features,bias=False)
self.Gaussian1 = Gaussian(h_features,out_features)
def forward(self,x):
x= self.linear(x)
print("The linear layer output is:",x)
x = self.Gaussian1(x)
return x
def Weight_init(m):
if isinstance(m,nn.Linear):
m.weight= nn.Parameter(torch.Tensor([[1,2,3],[4,5,6]]))
def Gaussian_init(m):
if isinstance(m, Gaussian):
m.sigma=nn.Parameter(torch.Tensor([3]))
m.center=nn.Parameter(torch.Tensor([[1,2]]))
model=RBF(3,2,1)
model.apply(Weight_init)
model.apply(Gaussian_init)
x=Variable(torch.Tensor([3,4,5]))
y=model(x)
print("The output of Gassian layer is :",y)
The output is always “0” ,even I changed the input several times.
And I still wonder if I need to consider the batch_size when defining own module
class RBF(nn.Module):
def __init__(self,in_features,h_features,out_features):
super(RBF,self).__init__()
self.linear = nn.Linear(in_features,h_features,bias=False)
self.Gaussian1 = Gaussian(h_features,out_features)
def forward(self,x):
x= self.linear(x)
#x = Gaussian(x)
self.Gaussian1(x)
return x
You should call self.Gaussian1 instead of constructor of Gaussian class, so as to pass x into Gaussian.forward()