Hello,
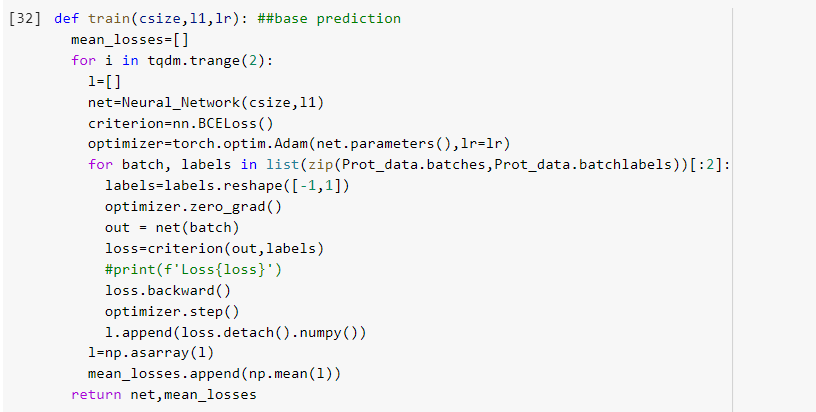
So my task is the following: I try to predict if an aminoacid is binding or not. The input for my network is a matrix of 22*9 meaining that there are 9 one hot encoded aminoacids.
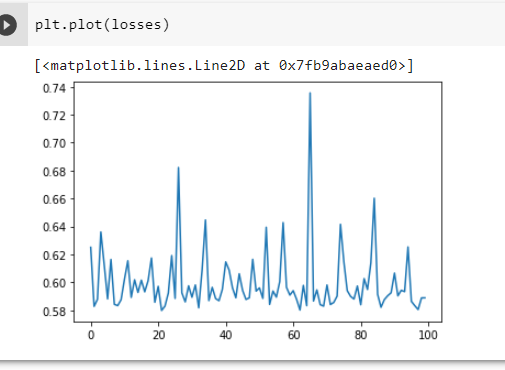
The problem is my network predicts pretty much the same value for every input. I tried changing layer size-> the value gets smaller but still pretty much uniform. Bigger or smaller learning rates did not help either. My loss per training iteration is jumping around and shows no order. The problem starts already in the first layer, where almost all values are close to 0, resulting in 0.5ish values after applying the sigmoid function. And so on. Backpropagation does not seem to work or something?
My code:
Here is an example of weights after one iteration(batchsize is 32 but 5 for demonstration purposes):
INPUT tensor([[0., 0., 0., …, 0., 0., 0.], ##One hot encoded aminoacids
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.]])
LAYER1 tensor([[-1.1884e-02, -1.3471e-01, -3.3829e-03, -2.1540e-01, 7.2217e-02,
-2.3076e-02, -1.5992e-01, 7.5262e-02, -2.7504e-03, -1.4549e-01,
3.3487e-04, -1.0971e-02, -2.2455e-01, 1.2630e-01, -1.7970e-02,
-1.2182e-01, 1.9458e-01, -1.0813e-01, -1.0313e-02, -6.6293e-02],
[-1.2023e-01, -1.4521e-01, 1.3482e-01, -1.9118e-01, 1.0324e-01,
-3.7693e-02, -2.2872e-01, 2.8561e-04, -4.9915e-02, -1.9751e-01,
1.9235e-02, -1.7252e-01, -6.9887e-02, 1.4740e-01, -1.1520e-01,
-3.3729e-01, -5.4954e-03, -4.1292e-02, -1.2781e-02, -3.1026e-02],
[-7.2280e-02, -9.4499e-02, 1.2107e-01, -1.1925e-01, -1.0798e-01,
2.7025e-01, -3.3732e-01, -9.9423e-02, -8.0310e-02, -2.7758e-03,
2.0783e-01, 5.7483e-04, 1.5967e-02, 1.4513e-01, -1.0606e-01,
8.6387e-02, -5.8707e-02, -2.6310e-02, -1.9591e-02, -2.7836e-01],
[-9.8013e-02, -1.6730e-01, -1.0512e-02, -3.8433e-02, -1.6118e-01,
1.8260e-01, -3.5972e-01, -3.3230e-02, 1.9475e-02, -9.6223e-02,
-4.7512e-02, -1.9144e-01, -3.6889e-02, -2.9930e-02, -2.1806e-01,
-5.8327e-02, -4.8562e-02, -1.0886e-01, 6.2563e-02, -1.2882e-01],
[-3.8484e-01, 4.3848e-02, 8.1674e-02, -2.8237e-01, -1.8457e-02,
-2.5487e-01, -3.0374e-01, -5.6237e-02, -6.0991e-02, 1.0755e-01,
3.8700e-02, -1.1368e-01, 1.3103e-01, -7.1677e-02, -3.4329e-02,
-2.5986e-01, 4.0630e-02, -1.9153e-02, 1.4287e-01, 2.2455e-02]],
grad_fn=)
SIG1 tensor([[0.4970, 0.4664, 0.4992, 0.4464, 0.5180, 0.4942, 0.4601, 0.5188, 0.4993,
0.4637, 0.5001, 0.4973, 0.4441, 0.5315, 0.4955, 0.4696, 0.5485, 0.4730,
0.4974, 0.4834],
[0.4700, 0.4638, 0.5337, 0.4524, 0.5258, 0.4906, 0.4431, 0.5001, 0.4875,
0.4508, 0.5048, 0.4570, 0.4825, 0.5368, 0.4712, 0.4165, 0.4986, 0.4897,
0.4968, 0.4922],
[0.4819, 0.4764, 0.5302, 0.4702, 0.4730, 0.5672, 0.4165, 0.4752, 0.4799,
0.4993, 0.5518, 0.5001, 0.5040, 0.5362, 0.4735, 0.5216, 0.4853, 0.4934,
0.4951, 0.4309],
[0.4755, 0.4583, 0.4974, 0.4904, 0.4598, 0.5455, 0.4110, 0.4917, 0.5049,
0.4760, 0.4881, 0.4523, 0.4908, 0.4925, 0.4457, 0.4854, 0.4879, 0.4728,
0.5156, 0.4678],
[0.4050, 0.5110, 0.5204, 0.4299, 0.4954, 0.4366, 0.4246, 0.4859, 0.4848,
0.5269, 0.5097, 0.4716, 0.5327, 0.4821, 0.4914, 0.4354, 0.5102, 0.4952,
0.5357, 0.5056]], grad_fn=)
OUT1 tensor([[0.6350],
[0.6277],
[0.6481],
[0.6043],
[0.6371]], grad_fn=)
SIG2 tensor([[0.6536],
[0.6520],
[0.6566],
[0.6466],
[0.6541]], grad_fn=)
BREAK
And after 20 training iterations:
ITERATION19
INPUT tensor([[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.]])
LAYER1 tensor([[-0.0629, 0.0192, -0.0056, -0.1177, -0.1336, 0.0947, 0.0761, 0.0111,
0.0353, 0.1887, -0.0601, 0.0876, -0.2136, -0.0462, -0.1947, 0.0695,
0.0689, -0.1898, 0.0435, 0.0465],
[-0.0374, -0.1245, -0.1563, 0.2059, 0.0215, 0.2642, -0.1232, -0.1641,
0.2543, 0.0636, -0.2663, 0.0397, -0.1072, -0.0468, -0.0068, 0.0056,
0.1969, -0.0082, -0.1935, -0.0064],
[-0.0047, -0.1265, -0.0111, 0.0955, 0.1450, -0.0402, -0.1283, -0.2326,
0.1164, -0.0493, -0.0772, -0.0174, 0.0515, -0.0974, -0.0123, -0.1615,
0.1239, 0.0990, 0.0468, 0.1425],
[ 0.1071, -0.0615, -0.0930, 0.0735, -0.2452, -0.1059, -0.0413, -0.0711,
0.4481, -0.1393, -0.2291, 0.0059, -0.2189, -0.1374, -0.0201, -0.0664,
0.2013, 0.0119, -0.0535, -0.1135],
[-0.0992, -0.1708, 0.0468, 0.0925, -0.1615, -0.0053, -0.0785, 0.0472,
0.1277, -0.0673, -0.2219, 0.0957, -0.0731, 0.0486, -0.0988, 0.1067,
0.1422, -0.0547, -0.0526, -0.0900]], grad_fn=)
100%|██████████| 20/20 [00:00<00:00, 24.03it/s]SIG1 tensor([[0.4843, 0.5048, 0.4986, 0.4706, 0.4666, 0.5237, 0.5190, 0.5028, 0.5088,
0.5470, 0.4850, 0.5219, 0.4468, 0.4885, 0.4515, 0.5174, 0.5172, 0.4527,
0.5109, 0.5116],
[0.4906, 0.4689, 0.4610, 0.5513, 0.5054, 0.5657, 0.4692, 0.4591, 0.5632,
0.5159, 0.4338, 0.5099, 0.4732, 0.4883, 0.4983, 0.5014, 0.5491, 0.4980,
0.4518, 0.4984],
[0.4988, 0.4684, 0.4972, 0.5239, 0.5362, 0.4899, 0.4680, 0.4421, 0.5291,
0.4877, 0.4807, 0.4956, 0.5129, 0.4757, 0.4969, 0.4597, 0.5309, 0.5247,
0.5117, 0.5356],
[0.5268, 0.4846, 0.4768, 0.5184, 0.4390, 0.4735, 0.4897, 0.4822, 0.6102,
0.4652, 0.4430, 0.5015, 0.4455, 0.4657, 0.4950, 0.4834, 0.5502, 0.5030,
0.4866, 0.4717],
[0.4752, 0.4574, 0.5117, 0.5231, 0.4597, 0.4987, 0.4804, 0.5118, 0.5319,
0.4832, 0.4448, 0.5239, 0.4817, 0.5121, 0.4753, 0.5267, 0.5355, 0.4863,
0.4869, 0.4775]], grad_fn=)
OUT1 tensor([[-0.0758],
[-0.0994],
[-0.0781],
[-0.0757],
[-0.0749]], grad_fn=)
SIG2 tensor([[0.4810],
[0.4752],
[0.4805],
[0.4811],
[0.4813]], grad_fn=)
BREAK
INPUT tensor([[0., 0., 0., …, 1., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.],
[0., 0., 0., …, 0., 0., 0.]])
LAYER1 tensor([[ 1.0191, -1.0846, -1.0535, 0.9865, 1.1097, 0.9978, 1.2395, -0.9887,
1.0245, 1.1841, 1.0048, 0.9626, -1.0941, 0.9735, 1.0480, 1.0463,
-0.7518, -0.9431, -0.9595, 0.8621],
[ 0.8312, -0.5470, -0.5932, 0.5750, 0.5846, 0.7568, 0.8730, -0.9507,
0.7150, 0.8542, 0.6390, 0.6484, -0.4502, 0.6243, 0.8885, 0.6466,
-0.7357, -0.8608, -0.7382, 0.6505],
[ 0.6720, -0.6638, -0.6548, 0.5123, 0.4758, 0.4278, 0.5499, -0.6849,
0.6195, 0.6485, 0.4727, 0.7233, -0.4381, 0.4723, 0.5971, 0.7836,
-0.5659, -0.6117, -0.6919, 0.6658],
[ 0.8300, -0.7890, -0.6093, 0.7198, 0.7867, 0.7110, 0.5804, -0.4863,
0.4393, 0.5117, 0.6362, 0.7359, -0.6294, 0.8104, 0.7539, 0.9311,
-0.5660, -0.7683, -0.8700, 0.7743],
[ 0.7943, -0.5147, -0.5071, 0.5075, 0.7360, 0.3048, 0.5949, -0.5193,
0.4593, 0.4101, 0.3451, 0.5035, -0.6488, 0.5498, 0.6608, 0.7725,
-0.4001, -0.4692, -0.2000, 0.5799]], grad_fn=)
SIG1 tensor([[0.7348, 0.2526, 0.2586, 0.7284, 0.7521, 0.7306, 0.7755, 0.2712, 0.7358,
0.7657, 0.7320, 0.7236, 0.2508, 0.7258, 0.7404, 0.7401, 0.3204, 0.2803,
0.2770, 0.7031],
[0.6966, 0.3666, 0.3559, 0.6399, 0.6421, 0.6806, 0.7054, 0.2787, 0.6715,
0.7015, 0.6545, 0.6567, 0.3893, 0.6512, 0.7086, 0.6562, 0.3239, 0.2972,
0.3234, 0.6571],
[0.6620, 0.3399, 0.3419, 0.6253, 0.6168, 0.6054, 0.6341, 0.3352, 0.6501,
0.6567, 0.6160, 0.6733, 0.3922, 0.6159, 0.6450, 0.6865, 0.3622, 0.3517,
0.3336, 0.6606],
[0.6964, 0.3124, 0.3522, 0.6726, 0.6871, 0.6706, 0.6412, 0.3808, 0.6081,
0.6252, 0.6539, 0.6761, 0.3476, 0.6922, 0.6800, 0.7173, 0.3622, 0.3168,
0.2953, 0.6844],
[0.6887, 0.3741, 0.3759, 0.6242, 0.6761, 0.5756, 0.6445, 0.3730, 0.6128,
0.6011, 0.5854, 0.6233, 0.3433, 0.6341, 0.6594, 0.6841, 0.4013, 0.3848,
0.4502, 0.6411]], grad_fn=)
OUT1 tensor([[-1.7504],
[-1.5769],
[-1.5207],
[-1.6025],
[-1.5091]], grad_fn=)
SIG2 tensor([[0.1480],
[0.1712],
[0.1794],
[0.1676],
[0.1811]], grad_fn=)