Hi,
I am building an OCR model in PyTorch and running into an issue with DataParallel and Packed RNN sequences.
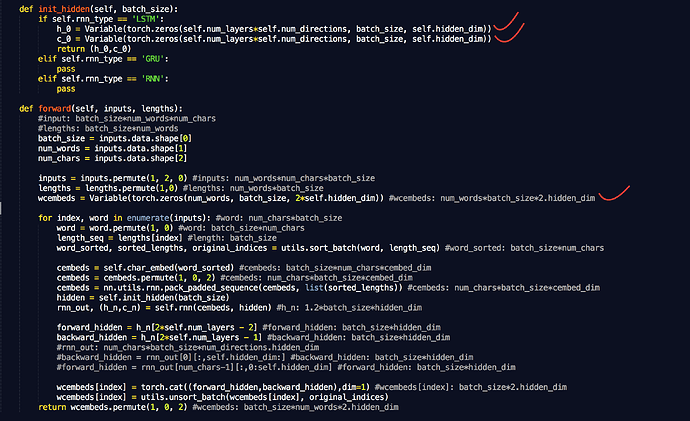
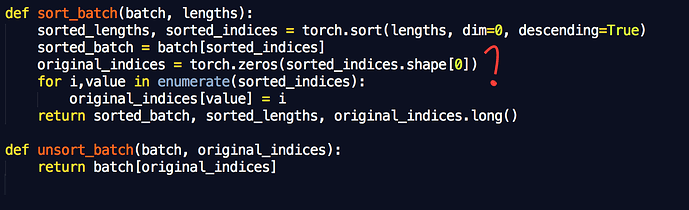
(1) My code seems pretty straightforward:
def forward(self, input, input_lengths):
packed_input = nn.utils.rnn.pack_padded_sequence(input, input_lengths)
packed_output = self.lstm(packed_input)
output, _ = nn.utils.rnn.pad_packed_sequence(packed_output)
return output
It works fine on a single GPU, but when I try to use DataParallel to run on multiple GPUs I run into a problem because the unpacked tensors on each GPU are of differing number of time steps, depending on the maximum value of the chunk of the input_lengths array that got passed to each GPU.
I am resolving the problem by doing this:
padded_output = Variable(torch.zeros(max_T, output.size()[1], output.size()[2]))
padded_output[:input_lengths.max(), :, :] = output
return padded_output
I just want to check with folks who are more familiar with PyTorch to see if this is the appropriate way to resolve this problem, or if there is a better way?
(2) Another issue, that is easily corrected by a small change to DataParallel, is that my model takes in an image tensor of dimension BCHW and outputs a set of LSTM outputs of dimension TBF – i.e. when I scatter inputs to the various GPUs I want to split on the batch dimension of my input vector, 0, and when I gather outputs from the various GPUs I want to stack them on the batch dimension of my output vector, 1.
I have modified DataParallel locally so that I can pass in different in_dim and out_dim parameters as opposed to a single dim parameter. Is there any interest in making such a change in the official repository?
Regards,
Stephen