Hi everyone!!
I am using this model architecture below to output a tensor with shape (1x313x64x64) where these 313 layers are probability distributions for each pixel of the image/tensor. As last layer, I am using a RELU to avoid negative values in the output.
##################################################
############## MODEL ARCHITECTURE ##############
##################################################
class Color_model(nn.Module):
def __init__(self):
super(Color_model, self).__init__()
self.features = nn.Sequential(
# conv1
nn.Conv2d(in_channels = 1, out_channels = 64, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 64, out_channels = 64, kernel_size = 3, stride = 2, padding = 1),
nn.ReLU(),
nn.BatchNorm2d(num_features = 64),
# conv2
nn.Conv2d(in_channels = 64, out_channels = 128, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 128, out_channels = 128, kernel_size = 3, stride = 2, padding = 1),
nn.ReLU(),
nn.BatchNorm2d(num_features = 128),
# conv3
nn.Conv2d(in_channels = 128, out_channels = 256, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 256, out_channels = 256, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 256, out_channels = 256, kernel_size = 3, stride = 2, padding = 1),
nn.ReLU(),
nn.BatchNorm2d(num_features = 256),
# conv4
nn.Conv2d(in_channels = 256, out_channels = 512, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 1),
nn.ReLU(),
nn.BatchNorm2d(num_features = 512),
# conv5
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.BatchNorm2d(num_features = 512),
# conv6
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 2, dilation = 2),
nn.ReLU(),
nn.BatchNorm2d(num_features = 512),
# conv7
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 1, dilation = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 1, dilation = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 512, out_channels = 512, kernel_size = 3, stride = 1, padding = 1, dilation = 1),
nn.ReLU(),
nn.BatchNorm2d(num_features = 512),
# conv8
nn.ConvTranspose2d(in_channels = 512, out_channels = 256, kernel_size = 4, stride = 2, padding = 1, dilation = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 256, out_channels = 256, kernel_size = 3, stride = 1, padding = 1, dilation = 1),
nn.ReLU(),
nn.Conv2d(in_channels = 256, out_channels = 256, kernel_size = 3, stride = 1, padding = 1, dilation = 1),
nn.ReLU(),
# conv8_313
nn.Conv2d(in_channels = 256, out_channels = 313, kernel_size = 1, stride = 1,dilation = 1),
nn.ReLU(),
# decoding
#nn.Conv2d(in_channels = 313, out_channels = 2, kernel_size = 1, stride = 1)
)
self.apply(weights_init)
#######################################
############## TEST CODE ##############
#######################################
import torch
from torch.autograd import Variable
from skimage.color import lab2rgb
from skimage.io import imread
from model import Color_model
#from data_loader import ValImageFolder
import numpy as np
from skimage.color import rgb2lab, rgb2gray
import torch.nn as nn
from PIL import Image
import scipy.misc
from scipy.misc import imsave
from torchvision import datasets, transforms
from training_layers import decode
import torch.nn.functional as F
import os
import imageio
scale_transform = transforms.Compose([
transforms.Resize((64,64),2),
#transforms.RandomCrop(224),
])
def load_image(image_path,transform=None):
rgb_image = Image.open(image_path)
if transform is not None:
rgb_image_resized = transform(rgb_image)
rgb_image_resized = np.asarray(rgb_image_resized)
lab_image_resized = rgb2lab(rgb_image_resized)
lab_image_resized = lab_image_resized.transpose(2,0,1)
img_l_resized = lab_image_resized[0,:,:]
img_l_resized = (np.round(img_l_resized)).astype(np.int) # L channel
img_l_resized = torch.from_numpy(img_l_resized).unsqueeze(0)
return img_l_resized
def main():
data_dir = "\\Dataset\\test\\images\\"
dirs=os.listdir(data_dir)
print(dirs)
color_model = Color_model().cuda().eval()
color_model.load_state_dict(torch.load('\\models\\model-1-1.ckpt'))
T = 0.38
soft = nn.Softmax2d()
Q_bins = np.load('\\code\\resources\\pts_in_hull.npy')
#print(Q_bins.shape)
#print(Q_bins)
for file in dirs:
img_l_resized = load_image(data_dir+'\\'+file, scale_transform)
img_l_resized = img_l_resized.unsqueeze(0).float().cuda()
img_ab_313 = color_model(img_l_resized)
img_ab_313 = img_ab_313.cpu()
img_ab_313_log_t = (torch.log10(img_ab_313))/T
print(img_ab_313_log_t.max())
soft_image_log_t = soft(img_ab_313_log_t)
When I get the output from the model (img_ab_313) and check min() and max() values, for the min() values there are some -inf values that I don’t understand.
I am using this formula (img_ab_313_log_t = (torch.log10(img_ab_313))/T) because this will be the input of softmax() function as described in an paper that I am trying to implement. See the formula below:
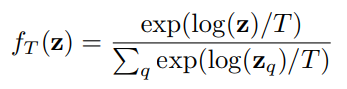
The torch.log10() don’t accept negative values because the result is nan…so I inserted the RELU in the end to avoid this…but I am getting -inf in the output…
Someone could help me with this? ![]()
Best regards,
Matheus Santos.


