@albanD, Hi, I have managed to reproduce it with a self-contained example adapted from distributed mnist
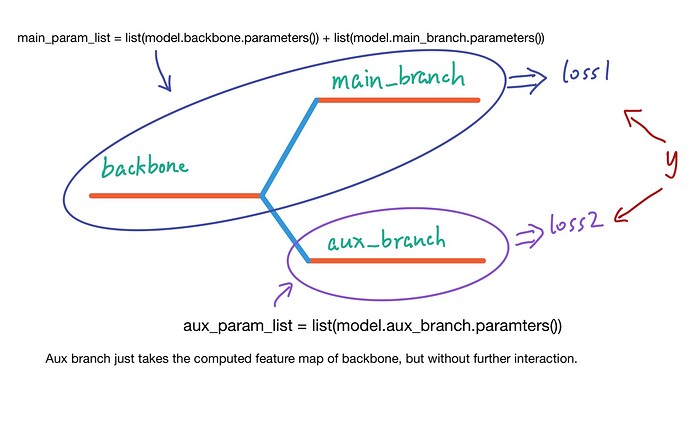
And I drew a sketch to illustrate my situation that may differ from the original one.
shown above, the aux branch’s loss wont affect the backbone. That’s what I think the “unused_parameters” means, But I dont know why it cant be done in DDP since it works fine in normal setting, also tested (you can try it out by comment the init_process_group part of code to disable the DDP)
then comes the code, it’s a bit of lengthy:grimacing:
from __future__ import division, print_function
import argparse
import torch
import torch.nn.functional as F
from torch import distributed, nn
from torch.utils import data
from torchvision import datasets, transforms
def distributed_is_initialized():
if distributed.is_available():
if distributed.is_initialized():
return True
return False
class Average(object):
def __init__(self):
self.sum = 0
self.count = 0
def __str__(self):
return '{:.6f}'.format(self.average)
@property
def average(self):
return self.sum / self.count
def update(self, value, number):
self.sum += value * number
self.count += number
class Accuracy(object):
def __init__(self):
self.correct = 0
self.count = 0
def __str__(self):
return '{:.2f}%'.format(self.accuracy * 100)
@property
def accuracy(self):
return self.correct / self.count
def update(self, output, target):
with torch.no_grad():
pred = output.argmax(dim=1)
correct = pred.eq(target).sum().item()
self.correct += correct
self.count += output.size(0)
class Trainer(object):
def __init__(self, model, optimizer1, optimizer2 , train_loader, test_loader, device):
self.model = model
self.optimizer1 = optimizer1
self.optimizer2 = optimizer2
self.train_loader = train_loader
self.test_loader = test_loader
self.device = device
def fit(self, epochs):
for epoch in range(1, epochs + 1):
train_loss, train_acc = self.train()
test_loss, test_acc = self.evaluate()
print(
'Epoch: {}/{},'.format(epoch, epochs),
'train loss: {}, train acc: {},'.format(train_loss, train_acc),
'test loss: {}, test acc: {}.'.format(test_loss, test_acc),
)
def train(self):
self.model.train()
train_loss = Average()
train_acc = Accuracy()
for data, target in self.train_loader:
data = data.to(self.device)
target = target.to(self.device)
output1, output2 = self.model(data)
loss1 = F.cross_entropy(output1, target)
self.optimizer1.zero_grad()
loss1.backward(retain_graph=True)
self.optimizer1.step()
loss2 = F.cross_entropy(output2, target)
self.optimizer2.zero_grad()
loss2.backward(retain_graph=True)
self.optimizer2.step()
train_loss.update(loss1.item(), data.size(0))
train_acc.update(output1, target)
return train_loss, train_acc
def evaluate(self):
self.model.eval()
test_loss = Average()
test_acc = Accuracy()
with torch.no_grad():
for data, target in self.test_loader:
data = data.to(self.device)
target = target.to(self.device)
output,_ = self.model(data)
loss = F.cross_entropy(output, target)
test_loss.update(loss.item(), data.size(0))
test_acc.update(output, target)
return test_loss, test_acc
class Net(nn.Module):
def __init__(self):
super(Net, self).__init__()
self.backbone = nn.Linear(784,64)
self.main_branch = nn.Linear(64, 10)
self.aux_branch = nn.Linear(64, 10)
def forward(self, x):
x = self.backbone(x.view(x.size(0), -1))
return self.main_branch(x), self.aux_branch(x)
class MNISTDataLoader(data.DataLoader):
def __init__(self, root, batch_size, train=True):
transform = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,)),
])
dataset = datasets.MNIST(root, train=train, transform=transform, download=True)
sampler = None
if train and distributed_is_initialized():
sampler = data.DistributedSampler(dataset)
super(MNISTDataLoader, self).__init__(
dataset,
batch_size=batch_size,
shuffle=(sampler is None),
sampler=sampler,
)
def run(args):
device = torch.device('cuda' if torch.cuda.is_available() and not args.no_cuda else 'cpu')
model = Net()
main_param_list = list(model.main_branch.parameters())+list(model.backbone.parameters())
aux_param_list = list(model.aux_branch.parameters())
optimizer1 = torch.optim.Adam(main_param_list, lr=args.learning_rate)
optimizer2 = torch.optim.Adam(aux_param_list, lr=args.learning_rate)
if distributed_is_initialized():
model.to(device)
model = nn.parallel.DistributedDataParallel(model)
else:
model = nn.DataParallel(model)
model.to(device)
train_loader = MNISTDataLoader(args.root, args.batch_size, train=True)
test_loader = MNISTDataLoader(args.root, args.batch_size, train=False)
trainer = Trainer(model, optimizer1,optimizer2, train_loader, test_loader, device)
trainer.fit(args.epochs)
def main():
parser = argparse.ArgumentParser()
parser.add_argument('--backend', type=str, default='nccl', help='Name of the backend to use.')
parser.add_argument(
'-i',
'--init-method',
type=str,
default='tcp://127.0.0.1:23456',
help='URL specifying how to initialize the package.')
parser.add_argument('-s', '--world-size', type=int, default=2, help='Number of processes participating in the job.')
parser.add_argument('-r', '--local_rank', type=int, default=0, help='Rank of the current process.')
parser.add_argument('--epochs', type=int, default=20)
parser.add_argument('--no-cuda', action='store_true')
parser.add_argument('-lr', '--learning-rate', type=float, default=1e-3)
parser.add_argument('--root', type=str, default='/mnt/EXTRA/remote/cosine/data')
parser.add_argument('--batch-size', type=int, default=128)
args = parser.parse_args()
print(args)
if args.world_size > 1:
distributed.init_process_group(
backend=args.backend,
init_method=args.init_method,
world_size=args.world_size,
rank=args.local_rank,
)
run(args)
if __name__ == '__main__':
main()
the error log is:
Traceback (most recent call last):
File "mnist.py", line 222, in <module>
main()
File "mnist.py", line 218, in main
run(args)
File "mnist.py", line 188, in run
trainer.fit(args.epochs)
File "mnist.py", line 71, in fit
train_loss, train_acc = self.train()
File "mnist.py", line 101, in train
loss2.backward(retain_graph=True)
File "/home/lxs/anaconda3/envs/torch10/lib/python3.6/site-packages/torch/tensor.py", line 107, in backward
torch.autograd.backward(self, gradient, retain_graph, create_graph)
File "/home/lxs/anaconda3/envs/torch10/lib/python3.6/site-packages/torch/autograd/__init__.py", line 93, in backward
allow_unreachable=True) # allow_unreachable flag
RuntimeError: has_marked_unused_parameters_ ASSERT FAILED at /opt/conda/conda-bld/pytorch_1556653183467/work/torch/csrc/distributed/c10d/reducer.cpp:181, please report a bug to PyTorch.
I am using two 1080TI on a single machine, using command python -m torch.distributed.launch --nproc_per_node=2 mnist.py to run it.
And I am using pytorch ver 1.1.0, and you may need to change the root path. I used the abs path.