I found the issue
this line of code was the problem, but I need to use this decomposition since with torch.svd sometimes I get error that the matrix is ill conditioned.
linalg.svd(X.cpu().detach().numpy(), full_matrices=False, lapack_driver=“gesvd”)
Yes, this line of code is causing the error since you are explicitly detaching the tensors from the computation graph and are calling into a 3rd party library (numpy in this case).
You would either have to use PyTorch operations or could write a custom autograd.Function using numpy operations and implement the backward pass manually as described here.
Thank you so much for your help.
Been struggling on this issue for a couple of hours. For me (1D CNN signal classification in Google Colab), it was as simple as:
torch.set_grad_enabled(True)
i have this error on my code
i have seached internet but i cant solve the problem
my code is:
def init_param(size,std=1):
return (torch.randn(size)*std).requires_grad_()
weight=init_param(784,1)
bias=init_param(1)
params=weight,bias
def train_epoch(lr,params):
for xb,yb in dltrain:
pred=(xb@weight+bias).view(-1,1)
pred=pred.sigmoid()
pred=(pred>.5).float()
loss=torch.where(yb==1,1-pred,pred).mean()#.requires_grad_()
weight.retain_grad()
bias.retain_grad()
loss.backward(retain_graph=True)
for p in weight,bias:
print(p.grad)
p.data-=(p.grad.data)*lr
p.grad=None
train_epoch(lr=0.5,params=params)
The following code is the source of your error -
You are essentially re-creating pred here which causes it to lose its computation graph.
It looks like a classification problem; try exploring loss functions like this one rather than defining your own like you’ve done here which also naturally has its requires_grad=False.
RuntimeError: element 0 of variables does not require grad and does not have a grad_fn here’s my training and it comes under loss.backward I would like to keep require grads To false as i don’t want them but I have to keep them on for model to work can you pls help
Hi,
Could you please post the code for the forward method of your custom model class Net?
Rather than screenshots you could include the code by enclosing between ```.
Also, apparently you are using the training_step from lightning - I don’t use lightning so not sure, but if there’s any code, please post.
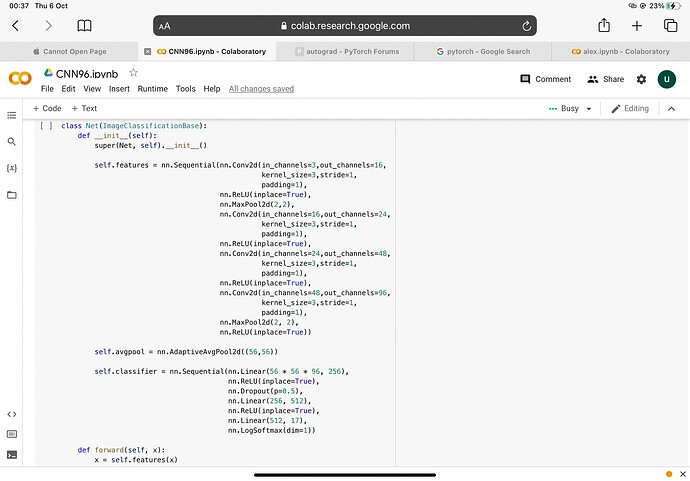
‘’'class ImageClassificationBase(nn.Module):
def training_step(self, batch):
images, labels = batch
out = self(images) # Generate predictions
loss = F.nll_loss(out, labels) # Calculate loss
return loss
def validation_step(self, batch):
images, labels = batch
out = self(images) # Generate predictions
loss = F.nll_loss(out, labels) # Calculate loss
acc = accuracy(out, labels) # Calculate accuracy
return {'val_loss': loss.detach(), 'val_acc': acc}
def validation_epoch_end(self, outputs):
batch_losses = [x['val_loss'] for x in outputs]
epoch_loss = torch.stack(batch_losses).mean() # Combine losses
batch_accs = [x['val_acc'] for x in outputs]
epoch_acc = torch.stack(batch_accs).mean() # Combine accuracies
return {'val_loss': epoch_loss.item(), 'val_acc': epoch_acc.item()}
def epoch_end(self, epoch, result):
print("Epoch [{}], train_loss: {:.4f}, val_loss: {:.4f}, val_acc: {:.4f}".format(
epoch, result['train_loss'], result['val_loss'], result['val_acc']))''''
then the model
‘’’ class Net(ImageClassificationBase):
def init(self):
super(Net, self).init()
self.features = nn.Sequential(nn.Conv2d(in_channels=3,out_channels=16,
kernel_size=3,stride=1,
padding=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(2,2),
nn.Conv2d(in_channels=16,out_channels=24,
kernel_size=3,stride=1,
padding=1),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=24,out_channels=48,
kernel_size=3,stride=1,
padding=1),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=48,out_channels=96,
kernel_size=3,stride=1,
padding=1),
nn.MaxPool2d(2, 2),
nn.ReLU(inplace=True))
self.avgpool = nn.AdaptiveAvgPool2d((56,56))
self.classifier = nn.Sequential(nn.Linear(56 * 56 * 96, 256),
nn.ReLU(inplace=True),
nn.Dropout(p=0.5),
nn.Linear(256, 512),
nn.ReLU(inplace=True),
nn.Linear(512, 17),
nn.LogSoftmax(dim=1))
def forward(self, x):
x = self.features(x)
x = self.avgpool(x)
x = x.view(-1, 56 * 56 * 96)
x = self.classifier(x)
return x ''''''
for param in model.parameters():
param.requires_grad = True
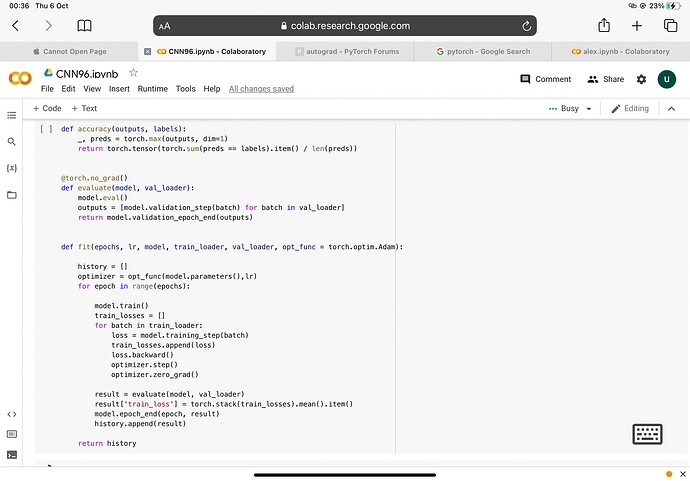
‘’'def accuracy(outputs, labels):
_, preds = torch.max(outputs, dim=1)
return torch.tensor(torch.sum(preds == labels).item() / len(preds))
@torch.no_grad()
def evaluate(model, val_loader):
model.eval()
outputs = [model.validation_step(batch) for batch in val_loader]
return model.validation_epoch_end(outputs)
def fit(epochs, lr, model, train_loader, val_loader, opt_func = torch.optim.Adam):
history = []
optimizer = opt_func(model.parameters(),lr)
for epoch in range(epochs):
model.train()
train_losses = []
for batch in train_loader:
loss = model.training_step(batch)
train_losses.append(loss)
loss.backward()
optimizer.step()
optimizer.zero_grad()
result = evaluate(model, val_loader)
result['train_loss'] = torch.stack(train_losses).mean().item()
model.epoch_end(epoch, result)
history.append(result)
return history
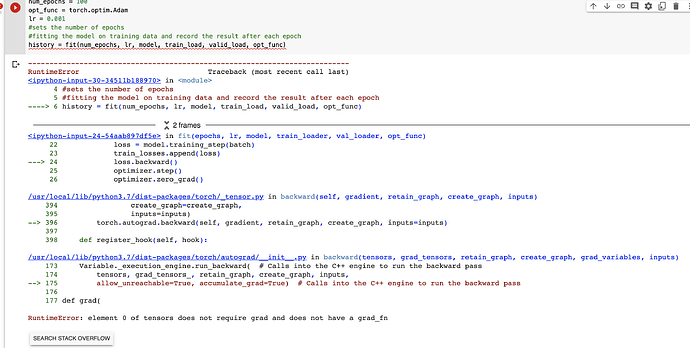
num_epochs = 100
opt_func = torch.optim.Adam
lr = 0.01
#sets the number of epochs
#fitting the model on training data and record the result after each epoch
history = fit(num_epochs, lr, model, train_load, valid_load, opt_func)
‘’‘’’
after few epochs the val loss keeps going up and training loss decreases and accuracy is 20-30% best, I want to turn the required gradient off in param_reuqire grads section but I keep running into runtime error element 0 does not require etc etc
Doesn’t this mean the model is already training and there’s no error when calling loss.backward() like you stated?
Anyway, please point me to the exact line where you face this error, as I didn’t quite understand this -
hello model runs when the Require grads are True but I want to run it with for param in model.parameters():
param.requires_grad = False
then it shows me to this error,
Yes, that’s expected.
Parameters are the leaf nodes and the ones with respect to which the gradient of the loss tensor is calculated. So, they need to have requires_grad=True.
Since you are already evaluating with torch.no_grad, there are no unnecessary gradient calculations taking place.
The loss problem has nothing to do with the parameters requiring gradients.
Try regularisation techniques.
Potentially, try learning rate schedulers and read about early stopping.
could you recommend me anything for this even based on the network and training parameters, my accuracy wont go above 33% Ideally 75/80% would be good. I really appreciate your help
This worked for me. Thanks.
I am trying to recreate CNN model from keras and facing runtime error on my loss.bakcward() (if I remove the function the model runs without learning)
I have check the model summary and saw summary was running fine with its channels and tensor, however due to my lack of knowledge I can not locate the bug. Can anyone give me an advice?
Then model :
from typing import List
class DNA_CNN_test2(nn.Module): # deepcre model
def __init__(self,
seq_len: int =1000,
#num_filters: List[int] = [64, 128, 64],
kernel_size: int = 8,
p = 0.25): # drop out value
super().__init__()
self.seq_len = seq_len
window_size = int(seq_len*(8/3000))
# CNN module
self.conv_net = nn.Sequential() # sequential containter. the forward() method of sequential accepts cany input and forwards it to yhe first module it contains
#num_filters = [4] + num_filters
self.model = nn.Sequential(
# conv block 1
nn.Conv1d(4,64,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.Conv1d(64,64,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.MaxPool1d(kernel_size=window_size),
nn.Dropout(p),
# conv block 2
nn.Conv1d(64,128,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.Conv1d(128,128,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.MaxPool1d(kernel_size=window_size),
nn.Dropout(p),
# conv block 3
nn.Conv1d(128,64,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.Conv1d(64,64,kernel_size=kernel_size, padding='same'),
nn.ReLU(inplace=True),
nn.MaxPool1d(kernel_size=window_size),
nn.Dropout(p),
nn.Flatten(),
nn.Linear(64*(seq_len//window_size**3), 1))
#nn.ReLU(inplace=True),
#nn.Dropout(p),
#nn.Linear(128, 64),
#nn.ReLU(inplace=True),
#nn.Linear(64*seq_len, 1))
def forward(self, xb: torch.Tensor):
"""Forward pass."""
# reshape view to batch_ssize x 4channel x seq_len
# permute to put channel in correct order
means (batch size, 4 channel - OHE(DNA), Seq.length )
xb = xb.permute(0, 2, 1).mean( dim = [1,2], keepdim = True).squeeze(dim= -1)
out = self.conv_net(xb)
return out
loss_batch,train and test step
# +--------------------------------+
# | Training and fitting functions |
# +--------------------------------+
def loss_batch(model, loss_func, xb, yb, opt=None,verbose=False):
'''
Apply loss function to a batch of inputs. If no optimizer
is provided, skip the back prop step.
'''
if verbose:
print('loss batch ****')
print("xb shape:",xb.shape)
print("yb shape:",yb.shape)
print("yb shape:",yb.squeeze(1).shape)
#print("yb",yb)
# get the batch output from the model given your input batch
# ** This is the model's prediction for the y labels! **
xb_out = model(xb.float())
if verbose:
print("model out pre loss", xb_out.shape)
#print('xb_out', xb_out)
print("xb_out:",xb_out.shape)
print("yb:",yb.shape)
print("yb.long:",yb.long().shape)
loss = loss_func(xb_out, yb.float()) # for MSE/regression
# __FOOTNOTE 2__
if opt is not None: # if opt
loss.backward()
opt.step()
opt.zero_grad()
return loss.item(), len(xb)
def train_step(model, train_dl, loss_func, device, opt):
'''
Execute 1 set of batched training within an epoch
'''
# Set model to Training mode
model.train()
tl = [] # train losses
ns = [] # batch sizes, n
# loop through train DataLoader
for xb, yb in train_dl:
# put on GPU
xb, yb = xb.to(device),yb.to(device)
# provide opt so backprop happens
t, n = loss_batch(model, loss_func, xb, yb, opt=opt)
# collect train loss and batch sizes
tl.append(t)
ns.append(n)
# average the losses over all batches
train_loss = np.sum(np.multiply(tl, ns)) / np.sum(ns)
return train_loss
def val_step(model, val_dl, loss_func, device):
'''
Execute 1 set of batched validation within an epoch
'''
# Set model to Evaluation mode
model.eval()
with torch.no_grad():
vl = [] # val losses
ns = [] # batch sizes, n
# loop through validation DataLoader
for xb, yb in val_dl:
# put on GPU
xb, yb = xb.to(device),yb.to(device)
# Do NOT provide opt here, so backprop does not happen
v, n = loss_batch(model, loss_func, xb, yb)
# collect val loss and batch sizes
vl.append(v)
ns.append(n)
# average the losses over all batches
val_loss = np.sum(np.multiply(vl, ns)) / np.sum(ns)
return val_loss
def fit(epochs, model, loss_func, opt, train_dl, val_dl,device,patience=1000):
'''
Fit the model params to the training data, eval on unseen data.
Loop for a number of epochs and keep train of train and val losses
along the way
'''
# keep track of losses
train_losses = []
val_losses = []
# loop through epochs
for epoch in range(epochs):
# take a training step
train_loss = train_step(model,train_dl,loss_func,device,opt)
train_losses.append(train_loss)
# take a validation step
val_loss = val_step(model,val_dl,loss_func,device)
val_losses.append(val_loss)
print(f"E{epoch} | train loss: {train_loss:.3f} | val loss: {val_loss:.3f}")
return train_losses, val_losses
def run_model(train_dl,val_dl,model,device,
lr=1e-2, epochs=50,
lossf=None,opt=None
):
'''
Given train and val DataLoaders and a NN model, fit the mode to the training
data. By default, use MSE loss and an SGD optimizer
'''
# define optimizer
if opt:
optimizer = opt
else: # if no opt provided, just use SGD
optimizer = torch.optim.SGD(model.parameters(), lr=lr)
# define loss function
if lossf:
loss_func = lossf
else: # if no loss function provided, just use MSE
loss_func = torch.nn.MSELoss()
# run the training loop
train_losses, val_losses = fit(
epochs,
model,
loss_func,
optimizer,
train_dl,
val_dl,
device)
return train_losses, val_losses
Error:
RuntimeError Traceback (most recent call last)
Cell In[51], line 5
2 DNA_CNN_test2 = DNA_CNN_test2(seq_len)
3 DNA_CNN_test2.to(device)
----> 5 DNA_CNN_test2_train_losses_lr4, DNA_CNN_test2_val_losses_lr4 = run_model(
6 train_dl,
7 val_dl,
8 DNA_CNN_test2,
9 device,
10 epochs=100,
11 lr= 1e-2
12 )
Cell In[42], line 139, in run_model(train_dl, val_dl, model, device, lr, epochs, lossf, opt)
136 loss_func = torch.nn.MSELoss()
138 # run the training loop
--> 139 train_losses, val_losses = fit(
140 epochs,
141 model,
142 loss_func,
143 optimizer,
144 train_dl,
145 val_dl,
146 device)
148 return train_losses, val_losses
Cell In[42], line 106, in fit(epochs, model, loss_func, opt, train_dl, val_dl, device, patience)
103 # loop through epochs
104 for epoch in range(epochs):
105 # take a training step
--> 106 train_loss = train_step(model,train_dl,loss_func,device,opt)
107 train_losses.append(train_loss)
109 # take a validation step
Cell In[42], line 54, in train_step(model, train_dl, loss_func, device, opt)
51 xb, yb = xb.to(device),yb.to(device)
53 # provide opt so backprop happens
---> 54 t, n = loss_batch(model, loss_func, xb, yb, opt=opt)
56 # collect train loss and batch sizes
57 tl.append(t)
Cell In[42], line 32, in loss_batch(model, loss_func, xb, yb, opt, verbose)
29 # __FOOTNOTE 2__
31 if opt is not None: # if opt
---> 32 loss.backward()
33 opt.step()
34 opt.zero_grad()
File /mnt/biostat/environments/parkj/dna2rna/lib/python3.11/site-packages/torch/_tensor.py:487, in Tensor.backward(self, gradient, retain_graph, create_graph, inputs)
477 if has_torch_function_unary(self):
478 return handle_torch_function(
479 Tensor.backward,
480 (self,),
(...)
485 inputs=inputs,
486 )
--> 487 torch.autograd.backward(
488 self, gradient, retain_graph, create_graph, inputs=inputs
489 )
File /mnt/biostat/environments/parkj/dna2rna/lib/python3.11/site-packages/torch/autograd/__init__.py:200, in backward(tensors, grad_tensors, retain_graph, create_graph, grad_variables, inputs)
195 retain_graph = create_graph
197 # The reason we repeat same the comment below is that
198 # some Python versions print out the first line of a multi-line function
199 # calls in the traceback and some print out the last line
--> 200 Variable._execution_engine.run_backward( # Calls into the C++ engine to run the backward pass
201 tensors, grad_tensors_, retain_graph, create_graph, inputs,
202 allow_unreachable=True, accumulate_grad=True)
RuntimeError: element 0 of tensors does not require grad and does not have a grad_fn
Based on your code self.conv_net is called in the forward method which is an empty nn.Sequential container. I’m not sure if the input will be directly returned in this case but if so, it would explain the error.
thank you for the reply. my input is written in specific way as it is DNA sequence and its corresponding value
class SeqDatasetOHE(Dataset):
'''
Dataset for one-hot-encoded sequences
'''
def __init__(self,
df,
seq_col='Seq',
target_col='boxcox_exp_mean'
):
# +--------------------+
# | Get the X examples |
# +--------------------+
# extract the DNA from the appropriate column in the df
self.seqs = list(df[seq_col].values)
self.seq_len = len(self.seqs[0])
# one-hot encode sequences, then stack in a torch tensor
self.ohe_seqs = torch.stack([torch.tensor(one_hot_encode(x)) for x in self.seqs])
# +------------------+
# | Get the Y labels |
# +------------------+
self.labels = torch.tensor(list(df[target_col].values)).unsqueeze(1)
def __len__(self): return len(self.seqs)
def __getitem__(self,idx):
# Given an index, return a tuple of an X with it's associated Y
# This is called inside DataLoader
seq = self.ohe_seqs[idx]
label = self.labels[idx]
return seq, label
#### buidling dataloader - batch size setting
currently batch size 4096 and it is fastai dataloader
## constructed DataLoaders from Datasets.
def build_dataloaders(train_df,
test_df,
seq_col='Seq',
target_col='boxcox_exp_mean',
batch_size=512,
shuffle=True
):
#Batch size – Refers to the number of samples in each batch.
#Shuffle – Whether you want the data to be reshuffled or not.
'''
Given a train and test df with some batch construction
details, put them into custom SeqDatasetOHE() objects.
Give the Datasets to the DataLoaders and return.
'''
# create Datasets
train_ds = SeqDatasetOHE(train_df,seq_col=seq_col,target_col=target_col)
test_ds = SeqDatasetOHE(test_df,seq_col=seq_col,target_col=target_col)
# Put DataSets into DataLoaders
train_dl = DataLoader(train_ds, batch_size=batch_size, shuffle=shuffle)
test_dl = DataLoader(test_ds, batch_size=batch_size)
return train_dl,test_dl
train_dl, val_dl = build_dataloaders(train_df, val_df)
Therefore I used xb.permute to arrange the data for the forward function. Would this be causing error?
No, permute won’t break the graph. Did you understand my concern in my previous post and did you check the forward method? It seems no trainable parameter or layers are used at all, since you are calling into self.conv_net not self.model.
thank you the relpy. Appolosize for missing understanding as I am new to the pytorch. I will try new model and will come back to the forum again!