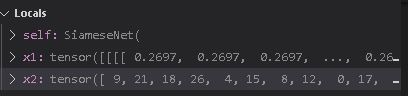
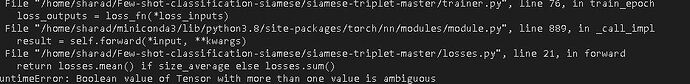
Thanks for the reply @ptrblck.
here is my classifier module which is reponsible for loading my dataset. The size of .img images that are in dataset are 1728*2393. I have such 21600 images in a single folder. I classified them using .csv file (Training,validation,testing).
ROOT_PATH = '/home/kumar/iter3/materials/'
Root_path1 = '/home/kumar/dataset/'
class Classifier(Dataset):
def __init__(self, setname,train=True):
csv_path = osp.join(ROOT_PATH, setname + '.csv')
lines = [x.strip() for x in open(csv_path, 'r').readlines()][1:]
data = []
label = []
lb = -1
self.wnids = []
for l in lines:
name, wnid = l.split(',')
path = osp.join(Root_path1, 'images', name)
if wnid not in self.wnids:
self.wnids.append(wnid)
lb += 1
data.append(path)
label.append(lb)
self.data = data
self.label = label
self.transform = transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(256),
transforms.ToTensor(),
#transforms.Normalize(mean=[0.485, 0.456, 0.406],
#std=[0.229, 0.224, 0.225])
transforms.Normalize(mean=[0.9439, 0.9439, 0.9439],
std=[0.208, 0.208, 0.208])
])
def __len__(self):
return len(self.data)
def __getitem__(self, i):
path, label = self.data[i], self.label[i]
image = self.transform(Image.open(path).convert('RGB'))
return image, label
below is my main module
#!/usr/bin/env python
# coding: utf-8
# In[ ]:
from __future__ import print_function, division
import torch
import torch.nn as nn
import torch.optim as optim
from torch.optim import lr_scheduler
import numpy as np
import torchvision
from torchvision import datasets, models, transforms
import matplotlib.pyplot as plt
import time
import os
import copy
from classifier import Classifier
plt.ion() # interactive mode
# In[ ]:
train_dataset = Classifier('train')
test_dataset = Classifier('val')
# In[ ]:
import torch
from torchvision.datasets import FashionMNIST
from torchvision import transforms
mean, std = 0.28604059698879553, 0.35302424451492237
batch_size = 256
cuda = torch.cuda.is_available()
kwargs = {'num_workers': 1, 'pin_memory': True} if cuda else {}
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size, shuffle=True, **kwargs)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size, shuffle=False, **kwargs)
n_classes = 27
# In[ ]:
import torch
from torch.optim import lr_scheduler
import torch.optim as optim
from torch.autograd import Variable
from trainer import fit
import numpy as np
cuda = torch.cuda.is_available()
'''
get_ipython().run_line_magic('matplotlib', 'inline')
import matplotlib
import matplotlib.pyplot as plt
classes = [................]
colors = ['#1f77b4', '#ff7f0e', '#2ca02c', '#d62728',
'#9467bd', '#8c564b', '#e377c2', '#7f7f7f',
'#bcbd22', '#17becf','#00FFFF','#7FFFD4',
'#F0FFFF','#F5F5DC','#0000FF','#A52A2A,',
'#7FFF00','#D2691E','#FF7F50','#006400',
'#FF00FF','#FFD700','#DAA520','#008000',
'#808080','#00FF00','#FFC0CB']
f_classes = classes
def plot_embeddings(embeddings, targets, xlim=None, ylim=None):
plt.figure(figsize=(10,10))
for i in range(10):
inds = np.where(targets==i)[0]
plt.scatter(embeddings[inds,0], embeddings[inds,1], alpha=0.5, color=colors[i])
if xlim:
plt.xlim(xlim[0], xlim[1])
if ylim:
plt.ylim(ylim[0], ylim[1])
plt.legend(f_classes)'''
def extract_embeddings(dataloader, model):
with torch.no_grad():
model.eval()
embeddings = np.zeros((len(dataloader.dataset), 2))
labels = np.zeros(len(dataloader.dataset))
k = 0
for images, target in dataloader:
if cuda:
images = images.cuda()
embeddings[k:k+len(images)] = model.get_embedding(images).data.cpu().numpy()
labels[k:k+len(images)] = target.numpy()
k += len(images)
return embeddings, labels
# In[ ]:
# Set up data loaders
batch_size = 256
kwargs = {'num_workers': 1, 'pin_memory': True} if cuda else {}
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size=batch_size, shuffle=True, **kwargs)
#for i_batch, sample_batched in enumerate(train_loader):
#print(i_batch, sample_batched)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size=batch_size, shuffle=False, **kwargs)
# Set up the network and training parameters
from networks import EmbeddingNet, ClassificationNet
from metrics import AccumulatedAccuracyMetric
embedding_net = EmbeddingNet()
model = ClassificationNet(embedding_net, n_classes=n_classes)
if cuda:
model.cuda()
loss_fn = torch.nn.NLLLoss()
lr = 1e-2
optimizer = optim.Adam(model.parameters(), lr=lr)
scheduler = lr_scheduler.StepLR(optimizer, 8, gamma=0.1, last_epoch=-1)
n_epochs = 100
log_interval = 10
# In[ ]:
#fit(train_loader, test_loader, model, loss_fn, optimizer, scheduler, n_epochs, cuda, log_interval, metrics=[AccumulatedAccuracyMetric()])
# In[ ]:
'''
train_embeddings_baseline, train_labels_baseline = extract_embeddings(train_loader, model)
plot_embeddings(train_embeddings_baseline, train_labels_baseline)
val_embeddings_baseline, val_labels_baseline = extract_embeddings(test_loader, model)
plot_embeddings(val_embeddings_baseline, val_labels_baseline)
'''
# In[ ]:
# Set up data loaders
from datasets import Siamese
# Step 1
siamese_train_dataset = Siamese(train_dataset) # Returns pairs of images and target same/different
siamese_test_dataset = Siamese(test_dataset)
batch_size = 128
kwargs = {'num_workers': 1, 'pin_memory': True} if cuda else {}
siamese_train_loader = torch.utils.data.DataLoader(siamese_train_dataset, batch_size=batch_size, shuffle=True, **kwargs)
siamese_test_loader = torch.utils.data.DataLoader(siamese_test_dataset, batch_size=batch_size, shuffle=False, **kwargs)
# Set up the network and training parameters
from networks import EmbeddingNet, SiameseNet
from losses import ContrastiveLoss
# Step 2
embedding_net = EmbeddingNet()
# Step 3
model = SiameseNet(embedding_net)
if cuda:
model.cuda()
# Step 4
margin = 1.
loss_fn = ContrastiveLoss(margin)
lr = 1e-3
optimizer = optim.Adam(model.parameters(), lr=lr)
scheduler = lr_scheduler.StepLR(optimizer, 8, gamma=0.1, last_epoch=-1)
n_epochs = 100
log_interval = 10
# In[ ]:
fit(siamese_train_loader, siamese_test_loader, model, loss_fn, optimizer, scheduler, n_epochs, cuda, log_interval)
# In[ ]:
'''
train_embeddings_cl, train_labels_cl = extract_embeddings(train_loader, model)
plot_embeddings(train_embeddings_cl, train_labels_cl)
val_embeddings_cl, val_labels_cl = extract_embeddings(test_loader, model)
plot_embeddings(val_embeddings_cl, val_labels_cl)
'''
This is my dataset.py module where I am defining my siamese function:
class Siamese(Dataset):
def __init__(self, dataset):
super().__init__()
self.dataset = dataset
def __getitem__(self, index):
# We need approx 50 % of images of the same class
same_class = random.randint(0, 1)
img_0, label_0 = self.dataset[index]
if same_class:
while True:
# keep looping till the same class image is found
index_1 = random.randint(0, self.__len__()-1)
img_1, label_1 = self.dataset[index_1]
if label_0 == label_1:
break
else:
while True:
index_1 = random.randint(0, self.__len__()-1)
img_1, label_1 = self.dataset[index_1]
if label_0 != label_1:
break
return (img_0, label_0), (img_1, label_1)
def __len__(self):
return len(self.dataset)
Here are all the losses function module:
import torch
import torch.nn as nn
import torch.nn.functional as F
class ContrastiveLoss(nn.Module):
"""
Contrastive loss
Takes embeddings of two samples and a target label == 1 if samples are from the same class and label == 0 otherwise
"""
def __init__(self, margin):
super(ContrastiveLoss, self).__init__()
self.margin = margin
self.eps = 1e-9
def forward(self, output1, output2, target, size_average=True):
distances = (output2 - output1).pow(2).sum(1) # squared distances
losses = 0.5 * (target.float() * distances +
(1 + -1 * target).float() * F.relu(self.margin - (distances + self.eps).sqrt()).pow(2))
return losses.mean() if size_average else losses.sum()
class TripletLoss(nn.Module):
"""
Triplet loss
Takes embeddings of an anchor sample, a positive sample and a negative sample
"""
def __init__(self, margin):
super(TripletLoss, self).__init__()
self.margin = margin
def forward(self, anchor, positive, negative, size_average=True):
distance_positive = (anchor - positive).pow(2).sum(1) # .pow(.5)
distance_negative = (anchor - negative).pow(2).sum(1) # .pow(.5)
losses = F.relu(distance_positive - distance_negative + self.margin)
return losses.mean() if size_average else losses.sum()
class OnlineContrastiveLoss(nn.Module):
"""
Online Contrastive loss
Takes a batch of embeddings and corresponding labels.
Pairs are generated using pair_selector object that take embeddings and targets and return indices of positive
and negative pairs
"""
def __init__(self, margin, pair_selector):
super(OnlineContrastiveLoss, self).__init__()
self.margin = margin
self.pair_selector = pair_selector
def forward(self, embeddings, target):
positive_pairs, negative_pairs = self.pair_selector.get_pairs(embeddings, target)
if embeddings.is_cuda:
positive_pairs = positive_pairs.cuda()
negative_pairs = negative_pairs.cuda()
positive_loss = (embeddings[positive_pairs[:, 0]] - embeddings[positive_pairs[:, 1]]).pow(2).sum(1)
negative_loss = F.relu(
self.margin - (embeddings[negative_pairs[:, 0]] - embeddings[negative_pairs[:, 1]]).pow(2).sum(
1).sqrt()).pow(2)
loss = torch.cat([positive_loss, negative_loss], dim=0)
return loss.mean()
class OnlineTripletLoss(nn.Module):
"""
Online Triplets loss
Takes a batch of embeddings and corresponding labels.
Triplets are generated using triplet_selector object that take embeddings and targets and return indices of
triplets
"""
def __init__(self, margin, triplet_selector):
super(OnlineTripletLoss, self).__init__()
self.margin = margin
self.triplet_selector = triplet_selector
def forward(self, embeddings, target):
triplets = self.triplet_selector.get_triplets(embeddings, target)
if embeddings.is_cuda:
triplets = triplets.cuda()
ap_distances = (embeddings[triplets[:, 0]] - embeddings[triplets[:, 1]]).pow(2).sum(1) # .pow(.5)
an_distances = (embeddings[triplets[:, 0]] - embeddings[triplets[:, 2]]).pow(2).sum(1) # .pow(.5)
losses = F.relu(ap_distances - an_distances + self.margin)
return losses.mean(), len(triplets)
metrics.py module:
import numpy as np
class Metric:
def __init__(self):
pass
def __call__(self, outputs, target, loss):
raise NotImplementedError
def reset(self):
raise NotImplementedError
def value(self):
raise NotImplementedError
def name(self):
raise NotImplementedError
class AccumulatedAccuracyMetric(Metric):
"""
Works with classification model
"""
def __init__(self):
self.correct = 0
self.total = 0
def __call__(self, outputs, target, loss):
pred = outputs[0].data.max(1, keepdim=True)[1]
self.correct += pred.eq(target[0].data.view_as(pred)).cpu().sum()
self.total += target[0].size(0)
return self.value()
def reset(self):
self.correct = 0
self.total = 0
def value(self):
return 100 * float(self.correct) / self.total
def name(self):
return 'Accuracy'
class AverageNonzeroTripletsMetric(Metric):
'''
Counts average number of nonzero triplets found in minibatches
'''
def __init__(self):
self.values = []
def __call__(self, outputs, target, loss):
self.values.append(loss[1])
return self.value()
def reset(self):
self.values = []
def value(self):
return np.mean(self.values)
def name(self):
return 'Average nonzero triplets'
networks.py module
import torch.nn as nn
import torch.nn.functional as F
class EmbeddingNet(nn.Module):
def __init__(self):
super(EmbeddingNet, self).__init__()
self.convnet = nn.Sequential(nn.Conv2d(3, 32, 5), nn.PReLU(),
nn.MaxPool2d(2, stride=2),
nn.Conv2d(32, 64, 5), nn.PReLU(),
nn.MaxPool2d(2, stride=2))
self.fc = nn.Sequential(nn.Linear(64 * 61 * 61, 256),
nn.PReLU(),
nn.Linear(256, 256),
nn.PReLU(),
nn.Linear(256, 2)
)
def forward(self, x):
output = self.convnet(x)
print('1:',output.shape)
output = output.view(output.size()[0], -1)
print('2:',output.shape)
output = self.fc(output)
output
print('3:',output.shape)
return output
def get_embedding(self, x):
return self.forward(x)
class EmbeddingNetL2(EmbeddingNet):
def __init__(self):
super(EmbeddingNetL2, self).__init__()
def forward(self, x):
output = super(EmbeddingNetL2, self).forward(x)
output /= output.pow(2).sum(1, keepdim=True).sqrt()
return output
def get_embedding(self, x):
return self.forward(x)
class ClassificationNet(nn.Module):
def __init__(self, embedding_net, n_classes):
super(ClassificationNet, self).__init__()
self.embedding_net = embedding_net
self.n_classes = n_classes
self.nonlinear = nn.PReLU()
self.fc1 = nn.Linear(2, n_classes)
def forward(self, x):
output = self.embedding_net(x)
output = self.nonlinear(output)
scores = F.log_softmax(self.fc1(output), dim=-1)
return scores
def get_embedding(self, x):
return self.nonlinear(self.embedding_net(x))
class SiameseNet(nn.Module):
def __init__(self, embedding_net):
super(SiameseNet, self).__init__()
self.embedding_net = embedding_net
def forward(self, x1, x2):
print('Sharad')
output1 = self.embedding_net(x1)
print('4:',output1.shape)
output2 = self.embedding_net(x2)
print('5:',output2.shape)
return output1, output2
def get_embedding(self, x):
return self.embedding_net(x)
class TripletNet(nn.Module):
def __init__(self, embedding_net):
super(TripletNet, self).__init__()
self.embedding_net = embedding_net
def forward(self, x1, x2, x3):
output1 = self.embedding_net(x1)
output2 = self.embedding_net(x2)
output3 = self.embedding_net(x3)
return output1, output2, output3
def get_embedding(self, x):
return self.embedding_net(x)
trainer.py module:
import torch
import numpy as np
def fit(train_loader, val_loader, model, loss_fn, optimizer, scheduler, n_epochs, cuda, log_interval, metrics=[],
start_epoch=0):
"""
Loaders, model, loss function and metrics should work together for a given task,
i.e. The model should be able to process data output of loaders,
loss function should process target output of loaders and outputs from the model
Examples: Classification: batch loader, classification model, NLL loss, accuracy metric
Siamese network: Siamese loader, siamese model, contrastive loss
Online triplet learning: batch loader, embedding model, online triplet loss
"""
for epoch in range(0, start_epoch):
scheduler.step()
for epoch in range(start_epoch, n_epochs):
scheduler.step()
# Train stage
train_loss, metrics = train_epoch(train_loader, model, loss_fn, optimizer, cuda, log_interval, metrics)
message = 'Epoch: {}/{}. Train set: Average loss: {:.4f}'.format(epoch + 1, n_epochs, train_loss)
for metric in metrics:
message += '\t{}: {}'.format(metric.name(), metric.value())
val_loss, metrics = test_epoch(val_loader, model, loss_fn, cuda, metrics)
val_loss /= len(val_loader)
message += '\nEpoch: {}/{}. Validation set: Average loss: {:.4f}'.format(epoch + 1, n_epochs,
val_loss)
for metric in metrics:
message += '\t{}: {}'.format(metric.name(), metric.value())
print(message)
def train_epoch(train_loader, model, loss_fn, optimizer, cuda, log_interval, metrics):
for metric in metrics:
metric.reset()
model.train()
losses = []
total_loss = 0
for batch_idx, (data, target) in enumerate(train_loader):
target = target if len(target) > 0 else None
if not type(data) in (tuple, list):
data = (data,)
if cuda:
data = tuple(d.cuda() for d in data)
if target is not None:
target = (t.cuda() for t in target)
optimizer.zero_grad()
outputs = model(*data)
if type(outputs) not in (tuple, list):
outputs = (outputs,)
loss_inputs = outputs
if target is not None:
target = (target,)
loss_inputs += target
loss_outputs = loss_fn(*loss_inputs)
loss = loss_outputs[0] if type(loss_outputs) in (tuple, list) else loss_outputs
losses.append(loss.item())
total_loss += loss.item()
loss.backward()
optimizer.step()
for metric in metrics:
metric(outputs, target, loss_outputs)
if batch_idx % log_interval == 0:
message = 'Train: [{}/{} ({:.0f}%)]\tLoss: {:.6f}'.format(
batch_idx * len(data[0]), len(train_loader.dataset),
100. * batch_idx / len(train_loader), np.mean(losses))
for metric in metrics:
message += '\t{}: {}'.format(metric.name(), metric.value())
print(message)
losses = []
total_loss /= (batch_idx + 1)
return total_loss, metrics
def test_epoch(val_loader, model, loss_fn, cuda, metrics):
with torch.no_grad():
for metric in metrics:
metric.reset()
model.eval()
val_loss = 0
for batch_idx, (data, target) in enumerate(val_loader):
target = target if len(target) > 0 else None
if not type(data) in (tuple, list):
data = (data,)
if cuda:
data = tuple(d.cuda() for d in data)
if target is not None:
target = target.cuda()
outputs = model(*data)
if type(outputs) not in (tuple, list):
outputs = (outputs,)
loss_inputs = outputs
if target is not None:
target = (target,)
loss_inputs += target
loss_outputs = loss_fn(*loss_inputs)
loss = loss_outputs[0] if type(loss_outputs) in (tuple, list) else loss_outputs
val_loss += loss.item()
for metric in metrics:
metric(outputs, target, loss_outputs)
return val_loss, metrics
utils.py
from itertools import combinations
import numpy as np
import torch
def pdist(vectors):
distance_matrix = -2 * vectors.mm(torch.t(vectors)) + vectors.pow(2).sum(dim=1).view(1, -1) + vectors.pow(2).sum(
dim=1).view(-1, 1)
return distance_matrix
class PairSelector:
"""
Implementation should return indices of positive pairs and negative pairs that will be passed to compute
Contrastive Loss
return positive_pairs, negative_pairs
"""
def __init__(self):
pass
def get_pairs(self, embeddings, labels):
raise NotImplementedError
class AllPositivePairSelector(PairSelector):
"""
Discards embeddings and generates all possible pairs given labels.
If balance is True, negative pairs are a random sample to match the number of positive samples
"""
def __init__(self, balance=True):
super(AllPositivePairSelector, self).__init__()
self.balance = balance
def get_pairs(self, embeddings, labels):
labels = labels.cpu().data.numpy()
all_pairs = np.array(list(combinations(range(len(labels)), 2)))
all_pairs = torch.LongTensor(all_pairs)
positive_pairs = all_pairs[(labels[all_pairs[:, 0]] == labels[all_pairs[:, 1]]).nonzero()]
negative_pairs = all_pairs[(labels[all_pairs[:, 0]] != labels[all_pairs[:, 1]]).nonzero()]
if self.balance:
negative_pairs = negative_pairs[torch.randperm(len(negative_pairs))[:len(positive_pairs)]]
return positive_pairs, negative_pairs
class HardNegativePairSelector(PairSelector):
"""
Creates all possible positive pairs. For negative pairs, pairs with smallest distance are taken into consideration,
matching the number of positive pairs.
"""
def __init__(self, cpu=True):
super(HardNegativePairSelector, self).__init__()
self.cpu = cpu
def get_pairs(self, embeddings, labels):
if self.cpu:
embeddings = embeddings.cpu()
distance_matrix = pdist(embeddings)
labels = labels.cpu().data.numpy()
all_pairs = np.array(list(combinations(range(len(labels)), 2)))
all_pairs = torch.LongTensor(all_pairs)
positive_pairs = all_pairs[(labels[all_pairs[:, 0]] == labels[all_pairs[:, 1]]).nonzero()]
negative_pairs = all_pairs[(labels[all_pairs[:, 0]] != labels[all_pairs[:, 1]]).nonzero()]
negative_distances = distance_matrix[negative_pairs[:, 0], negative_pairs[:, 1]]
negative_distances = negative_distances.cpu().data.numpy()
top_negatives = np.argpartition(negative_distances, len(positive_pairs))[:len(positive_pairs)]
top_negative_pairs = negative_pairs[torch.LongTensor(top_negatives)]
return positive_pairs, top_negative_pairs
class TripletSelector:
"""
Implementation should return indices of anchors, positive and negative samples
return np array of shape [N_triplets x 3]
"""
def __init__(self):
pass
def get_triplets(self, embeddings, labels):
raise NotImplementedError
class AllTripletSelector(TripletSelector):
"""
Returns all possible triplets
May be impractical in most cases
"""
def __init__(self):
super(AllTripletSelector, self).__init__()
def get_triplets(self, embeddings, labels):
labels = labels.cpu().data.numpy()
triplets = []
for label in set(labels):
label_mask = (labels == label)
label_indices = np.where(label_mask)[0]
if len(label_indices) < 2:
continue
negative_indices = np.where(np.logical_not(label_mask))[0]
anchor_positives = list(combinations(label_indices, 2)) # All anchor-positive pairs
# Add all negatives for all positive pairs
temp_triplets = [[anchor_positive[0], anchor_positive[1], neg_ind] for anchor_positive in anchor_positives
for neg_ind in negative_indices]
triplets += temp_triplets
return torch.LongTensor(np.array(triplets))
def hardest_negative(loss_values):
hard_negative = np.argmax(loss_values)
return hard_negative if loss_values[hard_negative] > 0 else None
def random_hard_negative(loss_values):
hard_negatives = np.where(loss_values > 0)[0]
return np.random.choice(hard_negatives) if len(hard_negatives) > 0 else None
def semihard_negative(loss_values, margin):
semihard_negatives = np.where(np.logical_and(loss_values < margin, loss_values > 0))[0]
return np.random.choice(semihard_negatives) if len(semihard_negatives) > 0 else None
class FunctionNegativeTripletSelector(TripletSelector):
"""
For each positive pair, takes the hardest negative sample (with the greatest triplet loss value) to create a triplet
Margin should match the margin used in triplet loss.
negative_selection_fn should take array of loss_values for a given anchor-positive pair and all negative samples
and return a negative index for that pair
"""
def __init__(self, margin, negative_selection_fn, cpu=True):
super(FunctionNegativeTripletSelector, self).__init__()
self.cpu = cpu
self.margin = margin
self.negative_selection_fn = negative_selection_fn
def get_triplets(self, embeddings, labels):
if self.cpu:
embeddings = embeddings.cpu()
distance_matrix = pdist(embeddings)
distance_matrix = distance_matrix.cpu()
labels = labels.cpu().data.numpy()
triplets = []
for label in set(labels):
label_mask = (labels == label)
label_indices = np.where(label_mask)[0]
if len(label_indices) < 2:
continue
negative_indices = np.where(np.logical_not(label_mask))[0]
anchor_positives = list(combinations(label_indices, 2)) # All anchor-positive pairs
anchor_positives = np.array(anchor_positives)
ap_distances = distance_matrix[anchor_positives[:, 0], anchor_positives[:, 1]]
for anchor_positive, ap_distance in zip(anchor_positives, ap_distances):
loss_values = ap_distance - distance_matrix[torch.LongTensor(np.array([anchor_positive[0]])), torch.LongTensor(negative_indices)] + self.margin
loss_values = loss_values.data.cpu().numpy()
hard_negative = self.negative_selection_fn(loss_values)
if hard_negative is not None:
hard_negative = negative_indices[hard_negative]
triplets.append([anchor_positive[0], anchor_positive[1], hard_negative])
if len(triplets) == 0:
triplets.append([anchor_positive[0], anchor_positive[1], negative_indices[0]])
triplets = np.array(triplets)
return torch.LongTensor(triplets)
def HardestNegativeTripletSelector(margin, cpu=False): return FunctionNegativeTripletSelector(margin=margin,
negative_selection_fn=hardest_negative,
cpu=cpu)
def RandomNegativeTripletSelector(margin, cpu=False): return FunctionNegativeTripletSelector(margin=margin,
negative_selection_fn=random_hard_negative,
cpu=cpu)
def SemihardNegativeTripletSelector(margin, cpu=False): return FunctionNegativeTripletSelector(margin=margin,
negative_selection_fn=lambda x: semihard_negative(x, margin),
cpu=cpu)
visualizing all embedding:
import os
import shutil
import numpy as np
import tensorflow as tf
import argparse
import json
from triplet_loss import batch_all_triplet_loss
from triplet_loss import batch_hard_triplet_loss
import mnist_dataset
from train_with_triplet_loss import my_model
from train_with_triplet_loss import test_input_fn
from tensorflow.contrib.tensorboard.plugins import projector
parser = argparse.ArgumentParser()
parser.add_argument('--data_dir', default='data',type=str, help="数据地址")
parser.add_argument('--model_dir', default='experiment/model', type=str, help="模型地址")
parser.add_argument('--model_config', default='experiment/params.json', type=str, help="模型参数")
parser.add_argument('--sprite_filename', default='experiment/mnist_10k_sprite.png', help="Sprite image for the projector")
parser.add_argument('--log_dir', default='experiment/log', type=str, help='可视化embeddings log文件夹')
def main(argv):
args = parser.parse_args(argv[1:])
'''创建模型'''
with open(args.model_config) as f:
params = json.load(f)
tf.logging.info("创建模型....")
config = tf.estimator.RunConfig(model_dir=args.model_dir, tf_random_seed=100) #
cls = tf.estimator.Estimator(model_fn=my_model, config=config, params=params) #
'''预测得到embeddings'''
tf.logging.info("预测....")
predictions = cls.predict(input_fn=lambda: test_input_fn(args.data_dir, params))
embeddings = np.zeros((10000, params['embedding_size']))
for i, p in enumerate(predictions):
embeddings[i] = p['embeddings']
tf.logging.info("embeddings shape: {}".format(embeddings.shape))
'''获得testset 的label 数据,并保存为metadata.tsv 文件'''
with tf.Session() as sess:
# Obtain the test labels
dataset = mnist_dataset.test(args.data_dir)
dataset = dataset.map(lambda img, lab: lab)
dataset = dataset.batch(10000)
labels_tensor = dataset.make_one_shot_iterator().get_next()
labels = sess.run(labels_tensor)
np.savetxt(os.path.join(args.log_dir, 'metadata.tsv'), labels, fmt='%d')
shutil.copy(args.sprite_filename, args.log_dir)
'''可视化embeddings'''
with tf.Session() as sess:
# 1. Variable
embedding_var = tf.Variable(embeddings, name="mnist_embeddings")
#tf.global_variables_initializer().run()
# 2.,embeddings.ckpt
saver = tf.train.Saver()
sess.run(embedding_var.initializer)
saver.save(sess, os.path.join(args.log_dir, 'embeddings.ckpt'))
# metadata.tsv, mnist_10k_sprite.png
summary_writer = tf.summary.FileWriter(args.log_dir)
config = projector.ProjectorConfig()
embedding = config.embeddings.add()
embedding.tensor_name = embedding_var.name
embedding.metadata_path = 'metadata.tsv'
embedding.sprite.image_path = 'mnist_10k_sprite.png'
embedding.sprite.single_image_dim.extend([28, 28])
projector.visualize_embeddings(summary_writer, config)
if __name__ == '__main__':
tf.reset_default_graph()
tf.logging.set_verbosity(tf.logging.INFO)
tf.app.run(main)
I am still wondering why I am getting this error. your help would be grateful.