Hi,
I’m trying to train an LSTM for language modeling using bigrams. I managed to get through the training (I don’t know if it was any good), but now I can’t use the model for inference.
Code for the dataset:
class RNNIndexLMDataset(data.Dataset):
def __init__(self, train):
self._bigrams = list(nltk.bigrams(train))
def __len__(self):
return len(self._bigrams)
def __getitem__(self, index):
w1, w2 = self._bigrams[index]
w1 = torch.LongTensor([w2i[w1]]).to(device)
w2 = torch.LongTensor([w2i[w2]]).to(device)
return w1, w2
Code for the model:
class RecurrentLM(nn.Module):
def __init__(self, vocab_size, embedding_dim, hidden_size, num_layers):
super(RecurrentLM, self).__init__()
self.embedding_dim = embedding_dim
self.hidden_size = hidden_size
self.num_layers = num_layers
self.embedding = nn.Embedding(vocab_size, embedding_dim)
self.lstm = nn.LSTM(embedding_dim, hidden_size, num_layers=self.num_layers)
self.fc = nn.Linear(hidden_size, vocab_size)
self.softmax = nn.LogSoftmax(dim=1)
def forward(self, input):
out = self.embedding(input)
hidden = self._init_hidden()
out = out.squeeze().unsqueeze(dim=0) # from (128, 1, 300) to (1, 128, 300) = (seqlen, batch, input)
out, hidden = self.lstm(out, hidden)
out = self.fc(out)
out = self.softmax(out)
return out
def _init_hidden(self):
return (torch.zeros(self.num_layers, BATCH_SIZE, self.hidden_size).to(device),
torch.zeros(self.num_layers, BATCH_SIZE, self.hidden_size).to(device))
Code for the training:
dataset = RNNIndexLMDataset(train)
data_loader = data.DataLoader(dataset, batch_size=BATCH_SIZE, shuffle=True, drop_last=True)
losses = []
loss_fn = nn.NLLLoss()
model = RecurrentLM(VOCAB_SIZE, EMBEDDING_DIM, NUM_HIDDEN, num_layers=1).cuda()
optimizer = optim.Adam(model.parameters())
epochs = 10
for _ in range(epochs):
total_loss = 0
for batch in data_loader:
w, y = batch
model.zero_grad()
yhat = model(w)
loss = loss_fn(yhat.squeeze(), y.squeeze())
loss.backward()
optimizer.step()
total_loss += loss
print('Loss: %.4f' % (total_loss.item()), end='\r')
losses.append(total_loss)
RNNLM = model
plt.plot(losses);
and finally, this is what crashes:
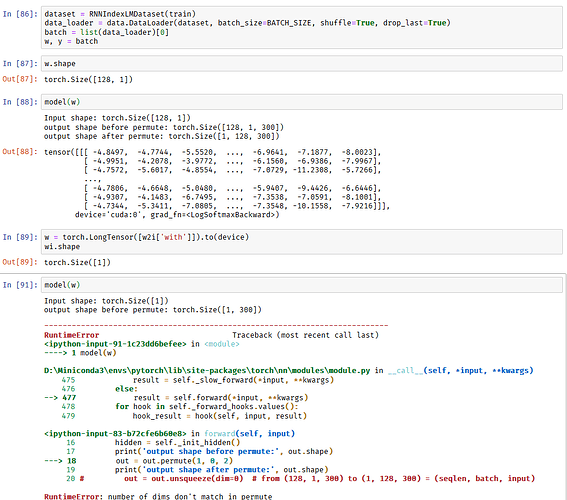
w = torch.LongTensor([[w2i['with']]]).to(device)
RNNLM(w.view(1, 1, -1))
Thanks!