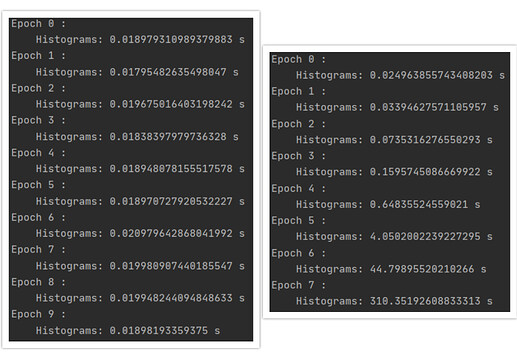
Hi Everyone,
I am using TensorBoard in my PyTorch project and I faced a problem. After each training epoch I log histograms of model parameters using SummaryWriter.add_histogram(). When I set weight_decay=0 in my optimizer everything works as expected (see left image below). However, when I set weight decay to some non-zero value, e.g. weight_decay=0.1, the histogram writing time starts to explode with every epoch (right image), rendering training impossible. Along with that, all my RAM is being filled.
Here is the code that demonstrates the problem:
import numpy as np
import time
import torch
import torch.nn as nn
import torch.nn.functional as F
from torch.utils.data import DataLoader
from torch.utils.data.dataloader import default_collate
from torch.utils.data.sampler import SubsetRandomSampler
from torchvision import datasets, transforms
from torch.utils.data import Subset
from torch.optim import Adam
from torch.optim.lr_scheduler import StepLR
from torch.utils.tensorboard import SummaryWriter
WEIGHT_DECAY = 0.1
EPOCHS = 10
class BaseDataLoader(DataLoader):
"""
Base class for all data loaders
"""
def __init__(self, dataset, batch_size, shuffle, validation_split, num_workers, collate_fn=default_collate):
self.validation_split = validation_split
self.shuffle = shuffle
self.batch_idx = 0
self.n_samples = len(dataset)
self.sampler, self.valid_sampler = self._split_sampler(self.validation_split)
self.init_kwargs = {
'dataset': dataset,
'batch_size': batch_size,
'shuffle': self.shuffle,
'collate_fn': collate_fn,
'num_workers': num_workers
}
super().__init__(sampler=self.sampler, **self.init_kwargs)
def _split_sampler(self, split):
if split == 0.0:
return None, None
idx_full = np.arange(self.n_samples)
np.random.seed(0)
np.random.shuffle(idx_full)
if isinstance(split, int):
assert split > 0
assert split < self.n_samples, "validation set size is configured to be larger than entire dataset."
len_valid = split
else:
len_valid = int(self.n_samples * split)
valid_idx = idx_full[0:len_valid]
train_idx = np.delete(idx_full, np.arange(0, len_valid))
train_sampler = SubsetRandomSampler(train_idx)
valid_sampler = SubsetRandomSampler(valid_idx)
# turn off shuffle option which is mutually exclusive with sampler
self.shuffle = False
self.n_samples = len(train_idx)
return train_sampler, valid_sampler
def split_validation(self):
if self.valid_sampler is None:
return None
else:
return DataLoader(sampler=self.valid_sampler, **self.init_kwargs)
class ADTargetTransform(object):
"""
Transform target labels to 0 for normal, 1 for anomaly.
"""
def __init__(self, normal_classes):
self.normal_classes = normal_classes
def __call__(self, target):
return int(target not in self.normal_classes)
class MnistADDataLoader(BaseDataLoader):
def __init__(self, data_dir, normal_classes, batch_size, oe_frac=0.0, shuffle=True, validation_split=0.0, num_workers=1, training=True):
# Create normal and outlier classes tuples
self.normal_classes = tuple(normal_classes)
self.outlier_classes = tuple([x for x in list(range(0, 10)) if x not in self.normal_classes])
trsfm = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.1307,), (0.3081,))
])
target_transform = ADTargetTransform(self.normal_classes)
self.data_dir = data_dir
mnist_dataset = datasets.MNIST(self.data_dir, train=training, download=True, transform=trsfm, target_transform=target_transform)
# Get indexes of normal and outlier classes
normal_idx = np.argwhere(np.isin(mnist_dataset.targets.cpu().data.numpy(), self.normal_classes)).flatten().tolist()
outlier_idx = np.argwhere(np.isin(mnist_dataset.targets.cpu().data.numpy(), self.outlier_classes)).flatten().tolist()
idx = normal_idx
if oe_frac > 0:
# Calculate how many outlier samples will be added to dataset
n_outliers_to_use = int(round(len(normal_idx)*oe_frac/(1-oe_frac)))
if n_outliers_to_use > len(outlier_idx):
n_outliers_to_use = len(outlier_idx)
idx += np.random.choice(outlier_idx, n_outliers_to_use).tolist()
np.random.shuffle(idx)
self.dataset = Subset(mnist_dataset, idx)
super().__init__(self.dataset, batch_size, shuffle, validation_split, num_workers)
class MnistModel(nn.Module):
def __init__(self, output_dim=10):
super().__init__()
self.conv1 = nn.Conv2d(1, 10, kernel_size=5)
self.conv2 = nn.Conv2d(10, 20, kernel_size=5)
self.conv2_drop = nn.Dropout2d()
self.fc1 = nn.Linear(320, 150)
self.fc2 = nn.Linear(150, 80)
self.fc3 = nn.Linear(80, output_dim)
def forward(self, x):
x = F.relu(F.max_pool2d(self.conv1(x), 2))
x = F.relu(F.max_pool2d(self.conv2_drop(self.conv2(x)), 2))
x = x.view(-1, 320)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
out = self.fc3(x)
return out
def loss_f(output, target):
eps = 1e-9
dists = torch.norm(output, p=2, dim=1) ** 2
losses = torch.where(target == 0, dists, -torch.log(1 - torch.exp(-dists) + eps))
return torch.mean(losses)
if __name__ == '__main__':
device = torch.device('cuda:0' if torch.cuda.is_available() else 'cpu')
data_loader = MnistADDataLoader(data_dir="data/", normal_classes=[0], batch_size=128,
oe_frac=0.1, shuffle=True, validation_split=0.1, num_workers=2)
valid_data_loader = data_loader.split_validation()
writer = SummaryWriter('log')
model = MnistModel(10).to(device)
trainable_params = filter(lambda p: p.requires_grad, model.parameters())
optimizer = Adam(trainable_params, lr=0.001, weight_decay=WEIGHT_DECAY, amsgrad=True)
lr_scheduler = StepLR(optimizer, step_size=50, gamma=0.1)
for epoch in range(0, EPOCHS):
print("Epoch", epoch, ":")
model.train()
for batch_idx, (data, target) in enumerate(data_loader):
data, target = data.to(device), target.to(device)
optimizer.zero_grad()
output = model(data)
loss = loss_f(output, target)
loss.backward()
optimizer.step()
start = time.time()
for name, p in model.named_parameters():
writer.add_histogram(name, p, bins='auto')
end = time.time()
print("\tHistograms:", end - start, "s")
I have killed quite some time to find the cause of my problem and a way to resolve it, however I still have no idea. I will be grateful if you can help me with that.
Thank you!