Hello, I am using crossentropyloss to segment CT scans. My data has 8 small structures of labels 1,2…8, and label 0 corresponds to areas that do not belong to the structures. In total, the structures make up only about 1.1% of the data, with the smallest structure being only about 0.001% and the largest around 0.9%. For testing purposes to debug weighting I am doing training where I set all the structures to a label of 1 and achieved reasonable results with no weights:
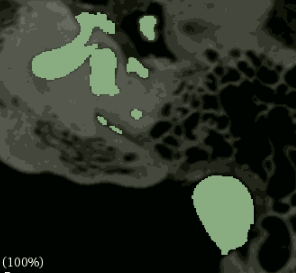
when compared to the input:
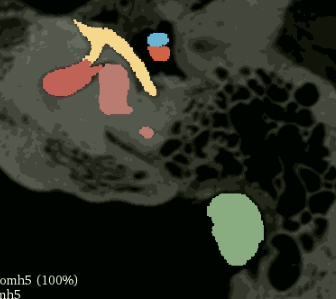
But I assumed that better results could be attained by adding weights to CrossEntropyLoss. I heard that using the inverse proportionality of labels was the correct weighting( in this case 0.9 for structures, 0.1 for non structures), but this severely overrepresented the structure labels:
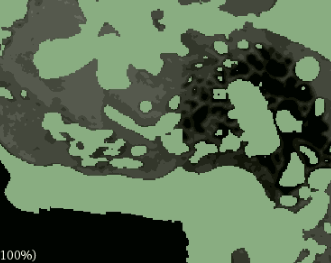
I trained again with less severe weights (0.8 for structures, 0.2 for non structures) and the pattern held. Overrepresentation of structures but less so.
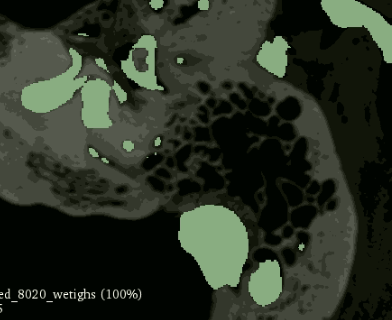
Any recommendations? Can I use any other techniques like weighted random samplers in segmentation, or is there a way I can make weights actually help my results?
Thanks, Kyle.