Hi there!
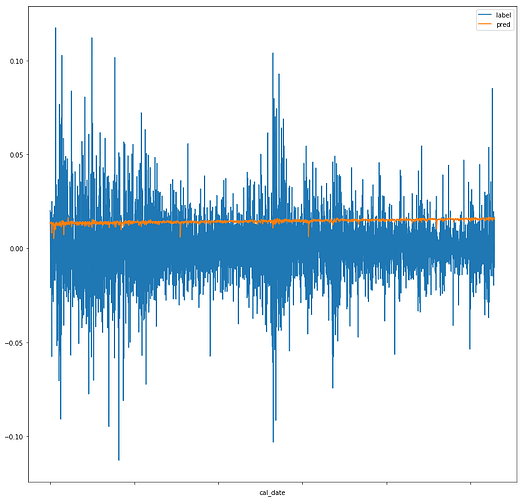
I’m working on a project which involves taking stock data and using the 10 previous days of stock data to predict the next day’s price (in the form of percent change). However, when I begin training the network, while my testing/validation losses do slowly decrease, they change by a fraction of a fraction of a percent. Attempting to use the network to generate predictions and then graphing the predictions vs the labels yields the following graph:
While the label fluctuates quite frequently, the pred value seems to stagnate, which is likely the cause of my losses not decreasing.
See below for details on the process used:
Input data:
-Data is made up of 495 stocks with data from their IPO or 1998 (whichever is earliest) to current
-Open, high, low, adj_close, and the label are all converted to pct_change from the previous day
-These columns are then scaled to a range of -1,1
-Date and volume are changed using the MinMaxScaler to be a value between 0,1
Sample of training data:
Network
class StockClassifier(nn.Module):
def __init__(self, input_length = 7,lstm_size = 64, lstm_layers=1, output_size = 1,
drop_prob=0.2):
super().__init__()
self.input_length = input_length
self.output_size = output_size
self.lstm_size = lstm_size
self.lstm_layers = lstm_layers
self.drop_prob = drop_prob
## TODO: define the LSTM
self.lstm = nn.LSTM(input_length, lstm_size, lstm_layers,
dropout=drop_prob, batch_first=False)
## TODO: define a dropout layer
self.dropout = nn.Dropout(drop_prob)
## TODO: define the final, fully-connected output layer
self.fc = nn.Linear(lstm_size, output_size)
def forward(self, nn_input, hidden_state):
'''
Perform a forward pass through the network
Args:
nn_input: the batch of input to NN
hidden_state: The LSTM hidden/cell state tuple
Returns:
logps: log softmax output
hidden_state: the updated hidden/cell state tuple
'''
# Input -> LSTM
lstm_out, hidden_state = self.lstm(nn_input, hidden_state)
# Stack up LSTM outputs -- this gets the final LSTM output for each sequence in the batch
lstm_out = lstm_out[-1, :, :]
# LSTM -> Dense Layer
lstm_out = self.dropout(self.fc(lstm_out))
# Return the final output and the hidden state
return lstm_out, hidden_state
def init_hidden(self, batch_size):
''' Initializes hidden state '''
# Create two new tensors with sizes n_layers x batch_size x n_hidden,
# initialized to zero, for hidden state and cell state of LSTM
weight = next(self.parameters()).data
hidden = (weight.new(self.lstm_layers, batch_size, self.lstm_size).zero_(),
weight.new(self.lstm_layers, batch_size, self.lstm_size).zero_())
return hidden
Training the network
criterion = nn.MSELoss()
optimizer = optim.SGD(model.parameters(), lr=learning_rate, momentum=momentum)
model.train()
training_losses = [x for x in range(epochs)]
validation_losses = [x for x in range(epochs)]
accuracies = [x for x in range(epochs)]
for epoch in range(epochs):
print('Starting Epoch {}'.format(epoch+1))
steps = 0
for t_batch, t_labels in dataloader(train_features, train_labels, batch_size=batch_size
,input_length=input_length, sequence_length=seq_length):
steps += 1
# Initialize Hidden/Cell state -- batch size is dynamic to account for batches that are not full
hidden = model.init_hidden(t_batch.shape[1])
hidden = tuple([each.data for each in hidden])
# Set tensors to correct device -- GPU or CPU
t_batch, t_labels = t_batch.to(device), t_labels.to(device)
for each in hidden:
each.to(device)
# Zero out gradients
optimizer.zero_grad()
# Run data through model -- output is output and new hidden/cell state
output, hidden = model(t_batch, hidden)
# Calculate loss and perform back prop -- clip grads if necessary
loss = criterion(output, t_labels)
loss.backward()
nn.utils.clip_grad_norm_(model.parameters(), clip)
# Take optimizer step
optimizer.step()
# VALIDATION OF MODEL#
if steps % print_every == 0:
model.eval()
val_losses = []
accuracy = []
#with torch.no_grad():
for val_batch, val_labels in dataloader(test_features, test_labels, batch_size=batch_size
,input_length=input_length, sequence_length=seq_length):
#Init hidden state -- again we have a dynamic batch size here
val_hidden = model.init_hidden(val_batch.shape[1])
val_hidden = tuple([each.data for each in val_hidden])
# Set device for tensors
val_batch, val_labels = val_batch.to(device), val_labels.to(device)
for each in val_hidden:
each.to(device)
# Run data through network
val_out, val_hidden = model(val_batch, val_hidden)
# Calculate and record loss
val_loss = criterion(val_out, val_labels)
val_losses.append(val_loss.item())
Using the trained model to generate the graph
seed = random.choice(tickers)
print("Testing for ticker: {}".format(seed))
stock_data = master_data.loc[master_data['ticker'] == seed, ['date', 'open', 'high', 'low', 'close', 'adj_close', 'volume', '50ma', 'label']]
#scale the data
stock_data.replace([np.inf, -np.inf], np.nan, inplace = True)
stock_data.dropna(inplace = True)
stock_data = scale_data(stock_data)
stock_data.replace([np.inf, -np.inf], np.nan, inplace = True)
stock_data.dropna(inplace = True)
#create graph data to compare label and pred values
graph_data = stock_data.filter(['date', 'label']).reset_index()
graph_data['pred'] = np.nan
#prepare the data for input to network
stock_data.drop(labels=['close', 'label'], inplace = True, axis = 1)
stock_data = stock_data.values
#have the network predict the 5 day prediction using sliding daily windows
i = 0
num_days = 10
model.eval()
while i <= len(stock_data)-num_days:
pred_data = torch.tensor(stock_data[i:i+num_days])
pred_data = pred_data.unsqueeze(1)
pred_data = pred_data.to(device)
pred_hidden = model.init_hidden(1)
pred_hidden = tuple([each.data for each in pred_hidden])
for each in pred_hidden:
each.to(device)
prediction, pred_hidden = model(pred_data, pred_hidden)
graph_data.loc[i+num_days-1, 'pred'] = prediction.item()
i += 1
graph_data.dropna(inplace = True)
graph_data
#graph both the label of the original value and the prediction value
ax = graph_data.plot.line(x = 'cal_date', y = ['label', 'pred'], figsize=(14,14))
plt.show()
Any help is appreciated! Thank you!
Jon