net is the instance of your model. In your case you should call model.apply(weight_init) 
thanks, Sorry to ask silly questions:)
Haha, please don’t be sorry for asking questions and feel free to ask again, in case you get stuck somewhere. 
Excuse me 
I am trying my trained networks on the test set which is patch of (11117)=847 , the model is defined as: model = ConvNetRedo1(32,64,(7,7,5),(5,5,3),500,100)
the input is the test patch with size of 847, why it is expected to have 32 1 7 7 5?!!
*** RuntimeError: Expected 5-dimensional input for 5-dimensional weight 32 1 7 7 5, but got 2-dimensional input of size [847, 1] instead
(Pdb) model
ConvNetRedo1(
(layer1): Sequential(
(0): Conv3d(1, 32, kernel_size=(7, 7, 5), stride=(1, 1, 1), padding=(2, 2, 2))
(1): ReLU()
(2): MaxPool3d(kernel_size=2, stride=2, padding=0, dilation=1, ceil_mode=False)
)
(layer2): Sequential(
(0): Conv3d(32, 64, kernel_size=(5, 5, 3), stride=(1, 1, 1), padding=(2, 2, 2))
(1): ReLU()
)
(fc1): Linear(in_features=5120, out_features=500, bias=True)
(drop_out1): Dropout(p=0.5, inplace=False)
(relu1): ReLU()
(fc2): Linear(in_features=500, out_features=100, bias=True)
(drop_out2): Dropout(p=0.5, inplace=False)
(relu2): ReLU()
(fc3): Linear(in_features=100, out_features=1, bias=True)
)
The first nn.Conv3d layer expects a 5-dimensional input with the shapes [batch_size, channels, depth, height, width], while you are apparently passing 1-dimensional input.
Many thanks for your answer.Yes because I want to test . I load my saved networks were CNN and now want to test on my patch which is 11117= 847 . ,the input should be 847 for testing . Am I wrong?
nn.Conv3d will not work with an input tensor of [11, 11, 7], as it’s expecting a 5-dimensional input.
I’m not sure which dimension represents what in your tensor, but you might want to add a channel and batch dimension to the tensor.
Once the dimensions are correct, you would have to make sure the input of the linear layer match the number of flattened activations.
I did data2=data1.reshape(1,11,11,7) which give me [1,1,11,11,7] for each patch to test. And it is working
sorry, how the pytorch initialize the weight basically, if it is not normal?
Parameters are initialized by default in the reset_parameters method of the corresponding layer.
sorry, it is outcome
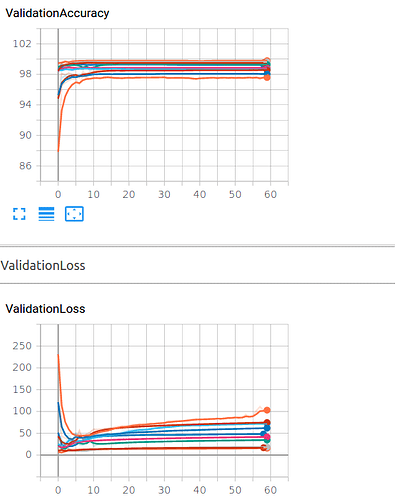
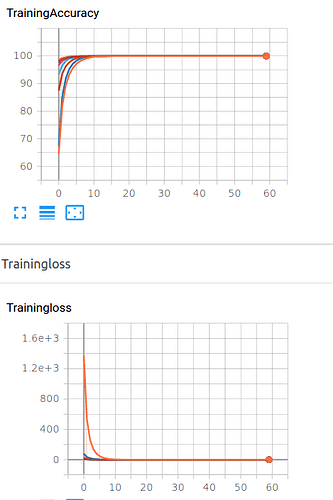
of my training for different iterations for 60 epoch in one fold. I think overfitting happen ,Do you agree?Hi Ptrblck
Sorry to take your time. I have a question if I want to find points location in 3 dimension from a MRI images , what is the command?
In Matlab I used (ind2sub) which used for 3 dimension.
np.unravel_index might be the numpy equivalent of the MATLAB function.
Side note: I’m not sure, if you are working with MATLAB arrays (and I’m not sure if you would run into this issue at all), but note that MATLAB uses column-major format, if I’m not mistaken. So you should test your indexing with some dummy data.
Sorry, I want to add label to my tensor board scalars in x and y axis (the x axis is epoch and y axis is Trainingloss). I start use tensor board with:
tb = SummaryWriter(root_dir3+’/Fold_’+str(FoldNum)+‘NumDarw=’+str(NumDraw)+str(Iteration))
and then I save the scalars by tb.add_scalar(‘Trainingloss’,LossScalar/Iterationnum,epoch+1)
Would you please tell me where I can add the label for x axis and y axis?
I’m not sure how to add labels to axes, but since your question is unrelated to the original topic, I would suggest to create a new thread so that more experienced users might see it.