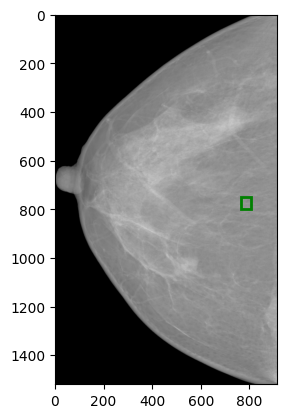
I have an original image of size (3518, 2800) as shown in the uploaded figure original_image.png. Now i have cropped the breast from the image and resized it to (1520, 912) as shown in the uploaded figure cropped_image.png. Following is the code for this preprocessing:
def np_CountUpContinuingOnes(b_arr):
# indice continuing zeros from left side.
# ex: [0,1,1,0,1,0,0,1,1,1,0] -> [0,0,0,3,3,5,6,6,6,6,10]
left = np.arange(len(b_arr))
left[b_arr > 0] = 0
left = np.maximum.accumulate(left)
# from right side.
# ex: [0,1,1,0,1,0,0,1,1,1,0] -> [0,3,3,3,5,5,6,10,10,10,10]
rev_arr = b_arr[::-1]
right = np.arange(len(rev_arr))
right[rev_arr > 0] = 0
right = np.maximum.accumulate(right)
right = len(rev_arr) - 1 - right[::-1]
return right - left - 1
def ExtractBreast(img):
img_copy = img.copy()
img = np.where(img <= 40, 0, img) # To detect backgrounds easily
height, _ = img.shape
# whether each col is non-constant or not
y_a = height // 2 + int(height * 0.4)
y_b = height // 2 - int(height * 0.4)
b_arr = img[y_b:y_a].std(axis=0) != 0
continuing_ones = np_CountUpContinuingOnes(b_arr)
# longest should be the breast
col_ind = np.where(continuing_ones == continuing_ones.max())[0]
img = img[:, col_ind]
# whether each row is non-constant or not
_, width = img.shape
x_a = width // 2 + int(width * 0.4)
x_b = width // 2 - int(width * 0.4)
b_arr = img[:, x_b:x_a].std(axis=1) != 0
continuing_ones = np_CountUpContinuingOnes(b_arr)
# longest should be the breast
row_ind = np.where(continuing_ones == continuing_ones.max())[0]
return img_copy[row_ind][:, col_ind]
def save_imgs(in_path, out_path, SIZE=(912, 1520)):
dicom = dicomsdl.open(in_path)
data = dicom.pixelData()
data = data[5:-5, 5:-5]
if dicom.getPixelDataInfo()['PhotometricInterpretation'] == "MONOCHROME1":
data = np.amax(data) - data
data = data - np.min(data)
data = data / np.max(data)
data = (data * 255).astype(np.uint8)
img = ExtractBreast(data)
img = cv2.resize(img, SIZE, interpolation=cv2.INTER_AREA)
cv2.imwrite(out_path, img)
print(out_path)
I am calling the save_imgs function to convert the dicom image of (3518, 2800) to png image of (1520, 912). Now my groundtruth bouding boxes are not adjusted by this transformation. I am using the code to transform the bounding box:
scale_x = 912 / 2800
scale_y = 1520 / 3518
# Resize the bounding box coordinates
resized_xmin = int(bounding_box[0] * scale_x)
resized_ymin = int(bounding_box[1] * scale_y)
resized_xmax = int(bounding_box[2] * scale_x)
resized_ymax = int(bounding_box[3] * scale_y)
can someone please help to transform the bounding box accordingly?
Here is original_image.png (extracted from dicom with size: (3518, 2800))
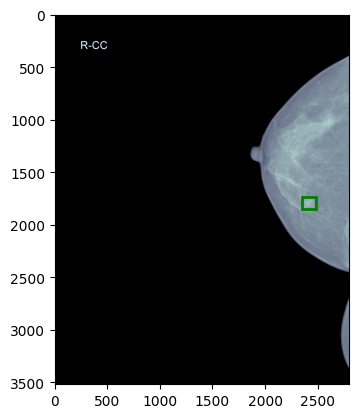
Here is cropped_image.png (1520, 912)