Hi all,
I am relatively new to Pytorch so please bare me with me ![]()
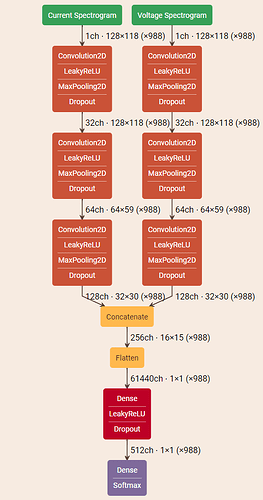
I have trained a network that takes as input 3 images. The net is based on this:
After training, I wish a heatmap for each input image for interpretability of results.
I make a prediction based on a triplet of images, and call backward(), but all my parameters and even my loss has no gradient:
pred.retain_grad()
pred[target_class].backward()
print(pred.grad) # gives None!
Here is the total print on the grad for model parameters:
trained_model.features_a4c.0.weight None
trained_model.features_a4c.0.bias None
trained_model.features_a4c.1.weight None
trained_model.features_a4c.1.bias None
trained_model.features_a4c.5.weight None
trained_model.features_a4c.5.bias None
trained_model.features_a4c.6.weight None
trained_model.features_a4c.6.bias None
trained_model.linear_block.0.weight None
trained_model.linear_block.0.bias None
trained_model.linear_block.3.weight None
trained_model.linear_block.3.bias None
linear_block.0.weight None
linear_block.0.bias None
linear_block.3.weight None
linear_block.3.bias None
model gradients = None
I have turned on model parameters requires_grad to True before backwards call, set model.eval().
Here is the gradcam model:
class MultiInputNet_GradCAM(nn.Module):
# Implement GradCAM hooks
def __init__(self, trained_model):
super().__init__() # inherit methods and attributes of nn.Module
self.trained_model = trained_model
self.features_a4c = self.trained_model.features_a4c[:8]
self.features_a2c = self.trained_model.features_a2c[:8]
self.features_a3c = self.trained_model.features_a3c[:8]
# we include the last maxpool2d and dropout in the forward function.
self.last_maxpool2d = nn.MaxPool2d(kernel_size=(2, 2), padding=1)
self.last_dropout = nn.Dropout(0.3)
self.gradients = None
self.linear_block = self.trained_model.linear_block
def activations_hook(self, grad):
self.gradients = grad
def forward(self, x_a4c, x_a2c, x_a3c):
x1 = self.features_a4c(x_a4c)
x2 = self.features_a2c(x_a2c)
x3 = self.features_a3c(x_a3c)
h1 = x1.register_hook(self.activations_hook)
h2 = x2.register_hook(self.activations_hook)
h3 = x3.register_hook(self.activations_hook)
x1 = self.last_maxpool2d(x1)
x1 = self.last_dropout(x1)
x2 = self.last_maxpool2d(x2)
x2 = self.last_dropout(x2)
x3 = self.last_maxpool2d(x3)
x3 = self.last_dropout(x3)
x_stack = torch.cat((x1, x2, x3), 1) # concatenate
x_stack = x_stack.view(x_stack.size(0), -1) # flatten batchwise (not fully), we want size of (batch_size, __)
out = self.linear_block(x_stack)
return out
# method for the gradient extraction
def get_gradient(self):
return self.gradients
# method for the activation extraction
def get_activations(self, x):
return self.features(x)
And finally (sorry for blocky codes :S ):
def my_gradcam(model, imgs, target_class):
model.cuda()
model.eval()
# get the most likely prediction of the model
pred = model(imgs[0].cuda(), imgs[1].cuda(), imgs[2].cuda()).argmax(dim=1)
pred=pred.float()
pred.requires_grad=True
pred.retain_grad()
pred[target_class].backward()
for name, param in model.named_parameters():
print(name, param.grad) # returns all None
# pull the gradients out of the model
gradients = model.get_gradient() # here self.gradients None too
Thank you in advance for your help. Any insight will be much appreciated!
Best,
David