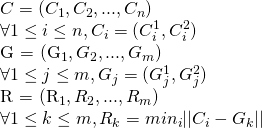def grid_minimal_dist(centroids,grids):
r"""
Get the minimal distance for each point
in the grid to a centroid
"""
# Number of sample
t = centroids.size()[0]
res = []
for c,g in zip(centroids,grids):
# Number of centroids
n = c.size()[0]
# Number of grid point
m = g.size()[0]
# Match the size of centroids and grid point
c = c.repeat(m,1)
g = g.repeat_interleave(n,dim=0)
# Compute the L2 norm
distance = g - c
distance = distance.pow(2)
distance = torch.sum(distance,1)
distance = torch.sqrt(distance)
# Regroup for each point of the grid
distance = distance.view(m,n)
# Get the minimal distance
distance,_ = torch.min(distance,1)
# Append the result
res.append(distance)
res = torch.stack(res)
return res
Let us note t the number of samples.
centroids is a collection (list, tuple, etc. ) of t matrix (n,2), n is the same as in the code
Example : centroids = [ [ [1,0], [0,2] ], [ [0,0] ] ]
grids is a (t,m,2) tensor. There are t grids of m points in a 2 dimensional space.
The purpose of this function is to compute the distance of the nearest centroids for each point in grids.
So, if we take 1 take centroid matrix [[[1,0],[0,2]]] and a grid [[[0,0]]].
In this example, k = 1, m = 1, n = 2
We want to compute the result vectorof length m (the size of the grid). The result in this case will be
res = [[1]]
Because the nearest point to (0,0) was (1,0) and their L2 norm is 1.
This is done inside the for loop.
Now I would like to parallelize this, so t can be as great as 1000 for example.
The only problem to using only element wise operation is because of n, as it may varies for each value of t. Therefore, centroids cannot be a tensor (unless by using a mask perhaps?)
Sorry if this is confusing. Please ask any question for clarification purposes.
Best,