Outline of the issue
I’ve made an autoencoder which takes in batches of tensors of shape (5,1,64,64). The model is trained to minimise the reconstruction loss as standard, but also the spread across the embedding values for each batch.
I’m using a fairly simple network as shown below with 3 convolutional layers and a single linearly connected embedding layer.
I’m finding that the embeddings are all converging to zero which I suspect is due to the “dying ReLU” problem as it is eleviated by using other activation functions e.g. tanh.
However, bizzarely, I’m seeing that despite the embeddings all having values of zero, the network is still able to reconstruct the input images to a good degree of accuracy.
Thoughts
It is my understanding that this shouldn’t be possible… Is there a way that a network can learn the identity function such that the embeddings can simply be zero’ed? To my knowledge, overfitting an autoencoder means that the network has a unique pathway for each input presented to the network, but still requires unique embedding values in order to reconstruct different inputs.
Any help on this weird behaviour would be greatly appreciated!
The model architecture
class CAE(torch.nn.Module):
def __init__(self):
super().__init__()
self.conv1 = torch.nn.Conv2d(1, 32, 3, padding=1)
self.conv2 = torch.nn.Conv2d(32, 64, 3, padding=1)
self.conv3 = torch.nn.Conv2d(64, 128, 3, padding=1)
self.lc1 = torch.nn.Linear(8 * 8 * 128, 6)
self.lc2 = torch.nn.Linear(6, 8 * 8 * 128)
self.trans1 = torch.nn.ConvTranspose2d(128, 64, 3, padding=1)
self.trans2 = torch.nn.ConvTranspose2d(64, 32, 3, padding=1)
self.trans3 = torch.nn.ConvTranspose2d(32, 1, 3, padding=1)
self.mp = torch.nn.MaxPool2d(2, return_indices=True)
self.up = torch.nn.MaxUnpool2d(2)
self.relu = torch.nn.ReLU()
def encoder(self, x):
x = self.conv1(x)
x = self.relu(x)
s1 = x.size()
x, ind1 = self.mp(x)
x = self.conv2(x)
x = self.relu(x)
s2 = x.size()
x, ind2 = self.mp(x)
x = self.conv3(x)
x = self.relu(x)
s3 = x.size()
x, ind3 = self.mp(x)
x = x.view(int(x.size()[0]), -1)
x = self.lc1(x)
x = self.relu(x)
return x, ind1, s1, ind2, s2, ind3, s3
def decoder(self, x, ind1, s1, ind2, s2, ind3, s3):
x = self.lc2(x)
x = x.view(int(x.size()[0]), 128, 8, 8)
x = self.up(x, ind3, output_size=s3)
x = self.relu(x)
x = self.trans1(x)
x = self.up(x, ind2, output_size=s2)
x = self.relu(x)
x = self.trans2(x)
x = self.up(x, ind1, output_size=s1)
x = self.relu(x)
x = self.trans3(x)
return x
def forward(self, x):
embeddings, ind1, s1, ind2, s2, ind3, s3 = self.encoder(x)
output = self.decoder(embeddings, ind1, s1, ind2, s2, ind3, s3)
return embeddings, output
Embeddings output during testing mode
Testing complete, compiling embeddings...
Embeddings compiled...
0 1 2 3 4 5
0 0.0 0.0 0.0 0.0 0.0 0.0
1 0.0 0.0 0.0 0.0 0.0 0.0
2 0.0 0.0 0.0 0.0 0.0 0.0
3 0.0 0.0 0.0 0.0 0.0 0.0
4 0.0 0.0 0.0 0.0 0.0 0.0
... ... ... ... ... ... ...
5600 0.0 0.0 0.0 0.0 0.0 0.0
5601 0.0 0.0 0.0 0.0 0.0 0.0
5602 0.0 0.0 0.0 0.0 0.0 0.0
5603 0.0 0.0 0.0 0.0 0.0 0.0
5604 0.0 0.0 0.0 0.0 0.0 0.0
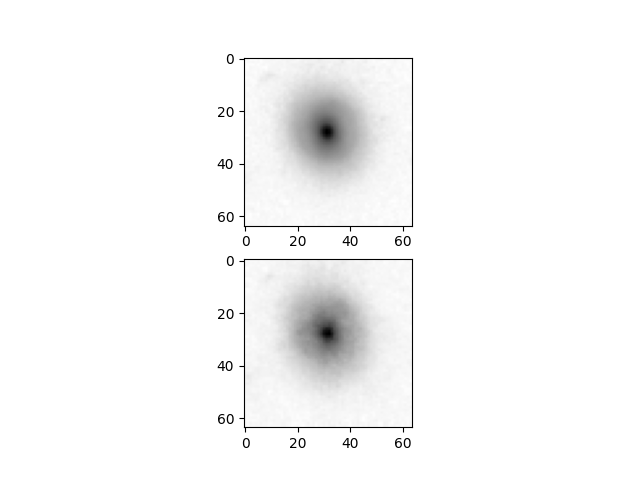
Example reconstruction image
Just to show that the model is indeed reconstructing the input images from the above embeddings (input=top, reconstruction=bottom):
The training procedure to check for data leakage
# =============================================================================
# Import packages
# =============================================================================
import matplotlib.pyplot as plt
import numpy as np
import os
import torch
from tqdm import tqdm
from utils.functions import learning_rate, training_plotter
# =============================================================================
# Define the training routine
# =============================================================================
def training(model:torch.nn.Module, device, dataset, indices, test_dataset, test_indices,
epochs, loss_function, initial_lr,plot_path=None,model_dir=None):
model = model.to(device).to(torch.float)
model.train(True)
print('Model moved to GPU...')
epoch_id, epoch_loss, epoch_std, train_test = [], [], [], np.zeros(epochs)
for epoch in tqdm(range(epochs)):
epoch_id.append(epoch)
running_loss = []
print("Epoch {0} of {1}" .format( (epoch+1), epochs))
optim = torch.optim.Adam(model.parameters(),learning_rate(initial_lr,epoch))
# =============================================================================
# Push data through the network
# =============================================================================
# =====================================================================
# Push the training data through every N epochs
# =====================================================================
for i in indices:
batch, _ = dataset[i]
# Stack the batch together and renormalise the image values to the range [-1,1]
batch = torch.stack(batch).to(device).to(torch.float) * 2 - 1
embeddings, prediction = model(batch)
# Minimise the difference between embeddings for a single batch
embedding_loss = torch.sum(torch.std(embeddings))
# Minimise the reconstruction loss
reconstruction_loss = loss_function(prediction, batch)
# Combine the two losses as one final loss for backprop
loss = reconstruction_loss + embedding_loss
running_loss.append(loss.detach().cpu())
optim.zero_grad(); loss.backward(); optim.step()
epoch_loss.append(np.mean(running_loss))
epoch_std.append(np.std(running_loss))
print("\n Mean loss: %.6f" % epoch_loss[epoch],
"\t Loss std: %.6f" % epoch_std[epoch],
"\t Learning rate: %.6f:" % learning_rate(initial_lr,epoch))
print('_'*73)
# =============================================================================
# Save the model
# =============================================================================
if (model_dir is not None) and (epoch == epochs-1):
torch.save(model.state_dict(),model_dir+'pilot_model.pt')
with…
# =============================================================================
# Setup learning rate decay function
# =============================================================================
def learning_rate(initial_lr, epoch):
"""Sets the learning rate to the initial LR decayed by a factor of 10 every
N epochs"""
lr = initial_lr * (0.99 ** (epoch// 1))
return lr