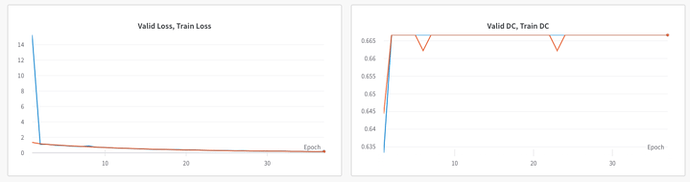
I am working with multi-class segmentation. I have 4 classes, my input to model has dimesnion : 32,1,384,384. The ground truth dimension is 32,4,384,384. The prediction from the model has the dimension 32,4,384,384. I am trying to calculate the loss using cross-entropy loss as : loss = CE_loss(preds, torch.argmax(var_gt, dim=1)) (I want to use this specific loss as I am replicating a paper and authors used it). However, the dice score remains constant for both train and val set after 2nd epoch as shown below:
.
To perform diagnostic, I followed Andrej Karpathy’s tip of overfitting the model with train data using 2 images. I did that, but my loss was not 0, it came out to be 1.6575. I am not able to figure out what is the issue here. Can anayone please guide me. Below is also the code for dice calcualtion, but I beleive the main problem here is loss.Below is given the code for how I am calculating the dice score:
eps = 0.0001
iflat = input_.reshape(-1)
tflat = target.reshape(-1)
if n_outputs == 1:
intersection = (iflat * tflat).sum()
union = iflat.sum() + tflat.sum()
dice = (2.0 * intersection + eps) / (union + eps)
if print_:
print(dice)
else:
dice = np.zeros(n_outputs-1)
for c in range(1,n_outputs): # assumes background is first class and doesn't compute its score
iflat_ = iflat==c
tflat_ = tflat==c
intersection = (iflat_ * tflat_).sum()
union = iflat_.sum() + tflat_.sum()
d = (2.0 * intersection + eps) / (union + eps)
if print_:
print(c, d)
dice[c-1] += d
if average:
dice = np.sum(dice)/(n_outputs-1)
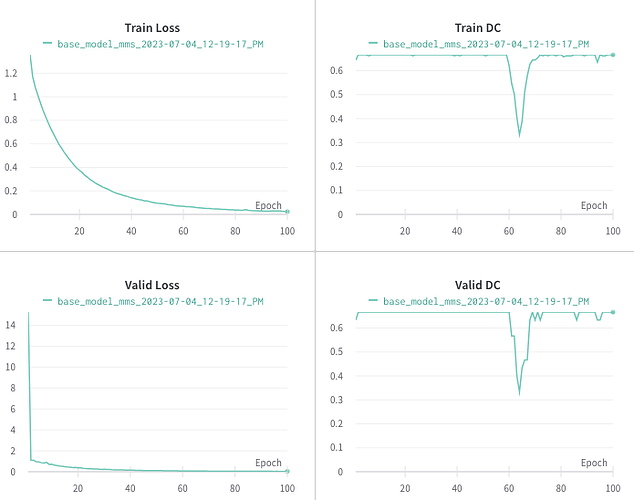
Since the new users can only put one embeded image i n the post, I am adding the results following Andrej’s tip here:
In your example code you are using some numpy operations, which could break the computation graph. Could you replace these using PyTorch operations and did you check the gradients in the model?
How dice calcualtion could break the computation graph? The graidents are updated on the basis of loss, while dice score is the evaluation critertion to save the best model checkpoint.
val_dice = dice_score(val_segmented_volume, gt_samples, n_outputs=n_channels_out)
if avg_val_dice > best_acc:
best_acc = avg_val_dice
print("best_acc- after updation", best_acc)
save_model(model, config, suffix, folder_time)
How to check the gradients in the model, I am not very expert in this field.
Using numpy would detach the tensor from the computation graph as seen here:
input_ = torch.randn(10, 10, requires_grad=True)
target = torch.randint(0, 2, (10, 10)).float()
n_outputs = input_.size(0)
eps = 0.0001
iflat = input_.reshape(-1)
tflat = target.reshape(-1)
dice = np.zeros(n_outputs-1)
for c in range(1,n_outputs): # assumes background is first class and doesn't compute its score
iflat_ = iflat==c
tflat_ = tflat==c
intersection = (iflat_ * tflat_).sum()
union = iflat_.sum() + tflat_.sum()
d = (2.0 * intersection + eps) / (union + eps)
print(c, d)
dice[c-1] += d
print(dice)
# [1.92307311e-06 1.00000000e+00 1.00000000e+00 1.00000000e+00
# 1.00000000e+00 1.00000000e+00 1.00000000e+00 1.00000000e+00
# 1.00000000e+00]
Even though input_ required gradients, the dice object is now a numpy array without any gradient history.
I guess you are recreating a tensor afterwards and set its .requires_grad attribute to True again as a workaround for this issue, which won’t reattach the tensor to the computation graph.
I would update numpy operation with torch. But for gradients I am still not clear what is your point. I am not updating the model based on dice score stats. I previously added only chunk for dice calculation, but before that in model.train() mode, I do calculate the loss and backpropagate the error based on that as follows:
for i, batch in enumerate(train_loader):
input_samples, gt_samples, voxel_dim = batch
if torch.cuda.is_available():
var_input = input_samples.to(device)
var_gt = gt_samples.to(device)
preds = model(var_input)
if n_channels_out == 1:
loss = weighted_cross_entropy_with_logits(preds, var_gt)
else:
loss = CE_loss(preds, torch.argmax(var_gt, dim=1))
train_loss_total += loss.item()
optimizer.zero_grad()
loss.backward()
optimizer.step()
Which means that in this step the gradients have been updated in the model. After that I caculate train dice. And to evaluate the model, I used validation set in model.eval() mode and calculated predictions, loss and dice score. And then saved the best checkpoint on the basis of dice score.
Thanks for clarifying. In that case I have misunderstood the question and don’t understand why you would expect to see a perfect dice score while overfitting two samples for a crossentropy loss.
Apologies for confuion! I was trying to debug the issue that is why I was overfitting the model with two samples to see weather the loss eventrually goes to 0. My main issue is that, the dice score remain constant to 0.667 after the second epoch for both train and val set as shown in
.
As per the learning curves model is learning, as loss is changing but it is not changing the dice accordingly. Should I be using softmax to un-normalized logits? Though the official documentation says that:
The input is expected to contain the unnormalized logits for each class (which do not need to be positive or sum to 1, in general).
You could visualize the predictions and try to estimate the dice as a quick smoke test.
The issue I’m seeing is that you expect to see a reduced dice loss without optimizing for it and assume your nn.CrossEntropyLoss is a good proxy for it.
Using a tiny subset of the data makes sense to make sure your model is able to overfit your currently used loss function, but I would not expect to see the same effect on other losses.
The same applies for e.g. nn.MSELoss. I would also not expect to see a perfect overfitting of it in your setup.