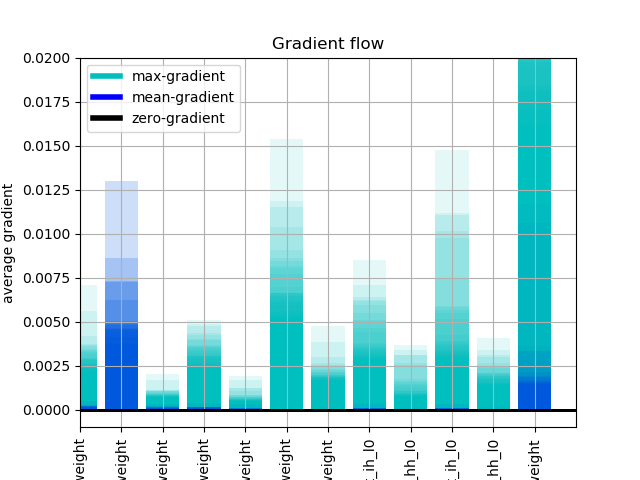
for completeness
I dont reccomend Tensorboard for many reasons, sure it is easy, but is a bit limited. But if you want these style graphs…
with SummaryWriter(log_dir=log_dir, comment="GradTest", flush_secs=30) as writer:
#... your learning loop
_limits = np.array([float(i) for i in range(len(gradmean))])
_num = len(gradmean)
writer.add_histogram_raw(tag=netname+"/abs_mean", min=0.0, max=0.3, num=_num,
sum=gradmean.sum(), sum_squares=np.power(gradmean, 2).sum(), bucket_limits=_limits,
bucket_counts=gradmean, global_step=global_step)
# where gradmean is np.abs(p.grad.clone().detach().cpu().numpy()).mean()
# _limits is the x axis, the layers
# and
_mean = {}
for i, name in enumerate(layers):
_mean[name] = gradmean[i]
writer.add_scalars(netname+"/abs_mean", _mean, global_step=global_step)