I am a novice at PyTorch, currently working on a school project involving the replication of a paper. I am currently creating a CNN to perform binary classification on medical documents (which I cannot share).
First, I activate CUDA, if possible:
device = (
"cuda"
if torch.cuda.is_available()
else "cpu"
)
print(f"Using {device} device")
I load my data like so.
study_corpus_tensor = torch.load("embedded_docs.pt")
study_corpus_tensor = study_corpus_tensor.to(device)
study_corpus_tensor is of size (number of documents, length of longest document, word embedding size). study_corpus_tensor[i, :, :] represents the i-th document. study_corpus_tensor[i, j, :] contains a word embedding (created using Word2Vec) for the j-th word in the i-th document.
This is my dataset class:
class CustomDatasetEmbedded(Dataset):
def __init__(self, corpus_tensor, labels):
self.x = corpus_tensor
self.y = labels
def __len__(self):
return len(self.y)
def __getitem__(self, index):
return (self.x[index, :, :], self.y[index])
I essentially pass in study_corpus_tensor to be stored as x and a column from a dataframe (1 if a patient had depression, 0 if they did not) to be stored as y.
depression_y = torch.tensor(labelled_corpus_df["Depression"]).to(device)
depression_dataset = CustomDatasetEmbedded(study_corpus_tensor, depression_y )
Here, I do a train test split. I do not have a validation set, as I am currently using the hyperparameters from the study.
train_dataset, test_dataset = torch.utils.data.random_split(depression_dataset, [0.8, 0.2])
train_loader = torch.utils.data.DataLoader(train_dataset, batch_size = 32, shuffle = True)
test_loader = torch.utils.data.DataLoader(test_dataset, batch_size = 32)
Here is my model. It is based on one of the models described in a paper:
class CNN_1_gram(nn.Module):
def __init__(self):
super(CNN_1_gram, self).__init__()
self.conv1 = nn.Conv2d(in_channels = 1,
out_channels = 100,
kernel_size = (1, embedding_vector_size),
stride = 1,
padding = 0)
# each kernel's feature map is condensed to a single value
conv1_output_height = study_corpus_tensor.shape[1] + 1 - 1
self.pool1 = nn.MaxPool2d(kernel_size = (conv1_output_height, 1))
self.do = nn.Dropout(p = 0.5)
self.fc= nn.Linear(100, 2) # Input size. 100, for 100 filters here.
self.activation = nn.LogSoftmax(dim = 1)
def forward(self, x):
# Provided Lua code (last_layer is probably output of pooling):
# local output = nn.LogSoftMax()(linear(nn.Dropout(opt.dropout_p)(last_layer)))
x = torch.unsqueeze(x, dim = 1)
x1 = self.conv1(x)
x1 = torch.relu(x1) # Point of this, given the global max pooling?
x = self.pool1(x1)
x = self.do(x)
x = torch.flatten(x, start_dim = 1)
x = self.fc(x)
x = self.activation(x)
return x
I instantiate the model:
cnn_1_gram_model = CNN_1_gram().to(device)
I create the optimizer and loss function, as described in the paper:
criterion = nn.modules.loss.NLLLoss()
optimizer = torch.optim.Adadelta(cnn_1_gram_model.parameters(), rho = 0.95, eps = 1e-6)
I create a training function:
n_epochs = 20
def train_model(model, train_dataloader, n_epoch, optimizer, criterion):
model.train()
for epoch in range(n_epoch):
curr_epoch_loss = []
for x, y in tqdm.tqdm(train_dataloader):
optimizer.zero_grad()
y_hat = model(x)
loss = criterion(y_hat, y)
loss.backward()
optimizer.step()
curr_epoch_loss.append(loss.cpu().data.numpy())
print(f"Epoch {epoch}: curr_epoch_loss={np.mean(curr_epoch_loss)}")
return model
I train the model (this is really fast):
cnn_1_gram_model = train_model(model = cnn_1_gram_model,
train_dataloader = train_loader,
n_epoch = n_epochs,
optimizer = optimizer,
criterion = criterion)
I create a function to evaluate the model:
def eval_model(model, dataloader):
model.eval()
Y_pred = []
Y_true = []
Y_score = []
with torch.no_grad():
for x, y in dataloader:
Y_true.append(y)
y_hat = model(x)
Y_score.append(y_hat[:, 1])
# Return class with higher probability
Y_pred.append(torch.max(y_hat, 1).indices)
Y_score = [y_score.to("cpu") for y_score in Y_score]
Y_pred = [y_pred.to("cpu") for y_pred in Y_pred]
Y_true = [y_true.to("cpu") for y_true in Y_true]
Y_score = np.concatenate(Y_score, axis = 0)
Y_pred = np.concatenate(Y_pred, axis=0)
Y_true = np.concatenate(Y_true, axis=0)
return Y_score, Y_pred, Y_true
I evaluate the model:
y_score, y_pred, y_true = eval_model(cnn_1_gram_model, test_loader)
I print some metrics:
print("Predicted percent of patients that have the condition:", np.sum(y_pred) / len(y_pred))
print("Actual percent of patients that have the condition:", np.sum(y_true) / len(y_true))
print("Accuracy:", accuracy_score(y_true, y_pred))
print("Precision:", precision_score(y_true, y_pred))
print("Recall:", recall_score(y_true, y_pred))
print("F1 Score:", f1_score(y_true, y_pred))
print("AUC:", roc_auc_score(y_true, y_score))
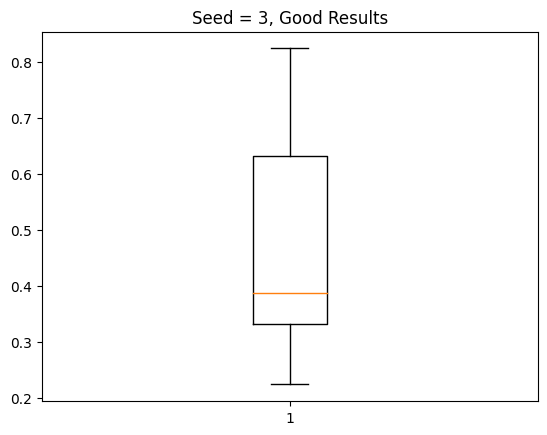
My results are extremely bizarre. About half the time, I get fairly good results like those seen below:
Predicted percent of patients that have the condition: 0.2462686567164179
Actual percent of patients that have the condition: 0.2947761194029851
Accuracy: 0.8917910447761194
Precision: 0.8787878787878788
Recall: 0.7341772151898734
F1 Score: 0.8
AUC: 0.9315518049695265
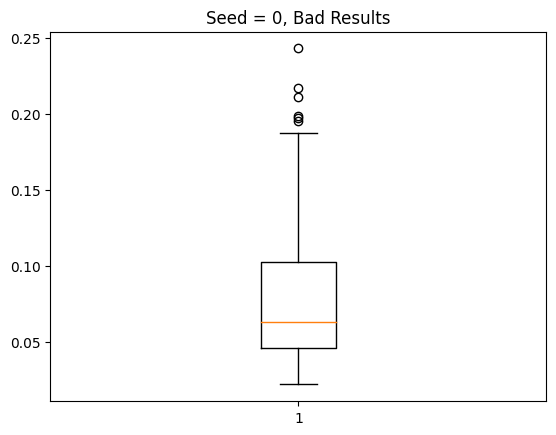
The other half of the time, I get abysmal results, with my model predicting class 0 for every single test example:
Predicted percent of patients that have the condition: 0.0
Actual percent of patients that have the condition: 0.26492537313432835
Accuracy: 0.7350746268656716
Precision: 0.0
Recall: 0.0
F1 Score: 0.0
AUC: 0.8657324658611568
/usr/local/lib/python3.9/dist-packages/sklearn/metrics/_classification.py:1344: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 due to no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
Still other times, I get nearly all 0s, with a precision of 1 and very low recall (indicating that the threshold to mark an example as positive seems to be extremely high).
I thought that differences in the division of data might be causing this issue, so I did:
for x, y in train_loader:
print(torch.sum(y))
and
for x, y in test_loader:
print(torch.sum(y))
but even when 0 was being outputted for every test example, there weren’t any batches without any positive instances in either the training or test dataset.
I found that when 0s are returned for every test instance, class 0 doesn’t have the highest probability for every training instance during the last training epoch. This, combined with the fact that training error always decreases as more and more epochs are completed, indicates that correct predictions are often being made during the training process. However, if I feed the training data into the evaluation function, it still returns 0 for all or nearly all of the training instances, which makes me think that overfitting isn’t the problem (as if there is overfitting, performance on the training data should be good).
This happens no matter how many epochs I choose (1, 5, or 20), and even when I change the loss and optimization functions. It happened both before and after I implemented GPU acceleration.
The authors of the study wrote that “After every parameter update, the parameters of the
feature maps were normalized to a norm of 3.” I talked with a TA about this, and he said it seemed strange and that I could ignore it. Did skipping this do something bad? I’m not sure if 3 is a good norm to use, as I may be using a different embedding size from the authors of the study (they did not record the size they used). How would you do this, anyways?
What might be going on? Did I make a stupid mistake somewhere? This problem seems extremely bizarre. The only thing that should be changing between runs is the division of data into training and test sets, and that should not lead to such large and confusing differences. Is something wrong with my eval_model() function?