Hi K. Frank!
Thanks so much for your response. Low level comments are super helpful because I’m at the stage where I’m mainly trying to discover/get used to what best practices are around for writing PyTorch code.
I don’t really understand your overall use case – “test utterance,” applying the model to a distance matrix – so I don’t have any thoughts on your “real” problem
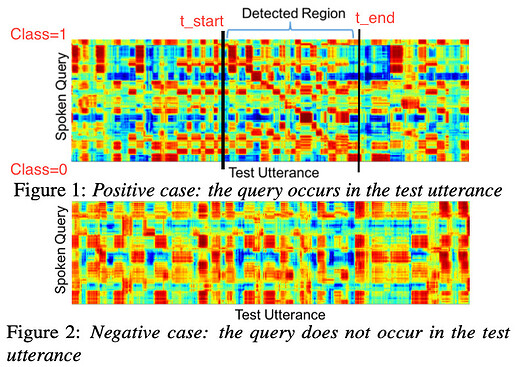
Ah yes, sorry, I didn’t explain it at all. The general task is called ‘query-by-example spoken term detection’ (QbE-STD) where you search for a spoken query (e.g. Q = ‘coffee’) in a corpus of audio documents (D1 = ‘I had coffee today’, D2 = ‘Where is the car?’) and output how likely the query is to occur in each document. If you don’t have access to a speech-to-text system, you typically just try to match on the spectral features (e.g. MFCCs) extracted from the audio files.
So the distance matrix is for a query Q of length M with F features and document D of length N with F features is a matrix of size M x N (taking the standardised Euclidean distance between each pair of feature vectors in Q and D). If the query occurs in the document, you typically get a quasi-diagonal band showing high spectrotemporal correlation somewhere along the document. I’m slowly making my way through replicating this paper while also learning PyTorch and looking at the GitHub repo associated with the paper.
While the approach in that paper is interesting, it occurred to me while learning about object detection in my deep learning class that I could try to extend this to the QbE-STD. My (naive) thought on this was to change the final layer from nn.Linear(60, 1) to nn.Linear(60, 3) (full model here) and make the network estimate, if a query occurs in the document, then at which time points (where t_start and t_end are proportion to the document, so in [0, 1]). Happy to hear any thoughts/cautions about this if you have any.
Assuming that a “class-0” labels row – that is, a row i for which labels[i, 0] is 0 – is all zeros – that is, for that i, labels[i, j] is 0.0 for all j – then simply using: MSE (output[:, -2:], labels[:, -2:]) (or MSE (output[:, 1:], labels[:, 1:]), see below), will provide that regularization. (Weight decay or a weight-regularization term in your loss would presumably provide equivalent benefit.)
Ah, I see. I don’t think I’ve grasped the entirety of what the loss function is doing then. I guess when I was playing around I was only looking at the output of MSE(), which is torch.Size([]). I’m guessing since MSE and BCE are instantiated as classes, e.g. MSE = torch.nn.MSELoss(), there are some internals in the object that keep track of the pairwise differences? So leaving the outputs as-is and doing the MSE will encourage the network to predict t_begin = 0 and t_end = 0 when class = 0, therefore constraining the weights that are associated with the 2nd and 3rd outputs when something like loss.backwards() is called? Please correct me if I’m wrong anywhere.
I was also wondering whether using 0 for values I wasn’t interested in was a good approach. One issue I see is confounding for t_start because for many cases where class = 1, the t_start is also 0, e.g. for ‘coffee’ in ‘Coffee’s the best!’ the t_start is 0 since the query ‘coffee’ is at the start of the document.
As a minor stylistic comment: Your choice of slicing notation looks rather perverse to me. For readability (at least for me), I would prefer, e.g., output[:, 0] to extract the first column, and output[:, 1:] to extract the remaining columns. output[:, :1] and output[:, 1:] could also work, as you could argue that using :1 and 1: together emphasizes that you are using all of the columns (but I still like 0 better for the first column).
Ah yes, this is the result of me trying various things in the console till the slice gave me what I wanted. Something I haven’t fully grasped is how : interacts with indices. I see suffixing the : to an index 1:, e.g. output[:, 1:], means return columns from 1 onwards. But when : is prefixed, as in output[:, :1], then the index starts from 1 instead of 0? So output[:, :2] returns the 1st and 2nd columns (not columns 0, 1, 2). For small label/output matrices, I think I might stick to explicit indexing with output[:, [0]] for the first column and use output[:, [0, 1]] for the 2nd and 3rd columns (coming from an R background).
Thanks!
Nay