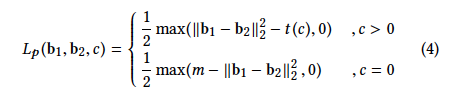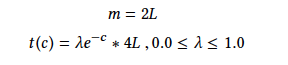Hi,
I am new in Pytorch and try to implement a custom loss function which is mentioned in a paper, Deep Multi-Similarity Hashing for Multi-label Image Retrieval.
However, the loss value from the loss function does not converge to smaller values whether used big epoch numbers or not. It plays around the started value.
I checked weight values and they are changing so I think backward mechanisms work.
So where is the problem?
The loss function:
class pairWiseLoss(nn.Module):
def forward(self, binaryCodes, labels):
lambdaValue = 0.62
numSample = len(binaryCodes)
halfNumSample = numSample // 2
similarity = torch.diagonal(torch.mm(labels[:halfNumSample], labels[halfNumSample:].t()))
maskCommonLabel = similarity.gt(0.0)
maskNoCommonLabel = similarity.eq(0.0)
tc = torch.exp(-similarity) * lambdaValue * 4 * args.bits
m = 2 * args.bits
hammingDistance = torch.diagonal( torch.cdist(binaryCodes[:halfNumSample],binaryCodes[halfNumSample:], p = 0) )
zeros = torch.zeros_like(hammingDistance)
loss = torch.sum(torch.masked_select(0.5 * torch.max(hammingDistance - tc , zeros ),maskCommonLabel ) ) + torch.sum(torch.masked_select(0.5 * torch.max( m - hammingDistance, zeros), maskNoCommonLabel ) )
return loss
The training block:
def train(trainloader, model, optimizer, epoch, use_cuda, train_writer,gpuDisabled,customLoss):
label_train = []
name_train = []
generated_codes = []
lossTracker = MetricTracker()
model.train()
for idx, data in enumerate(tqdm(trainloader, desc="training")):
numSample = data["bands10"].size(0)
if gpuDisabled :
bands = torch.cat((data["bands10"], data["bands20"],data["bands60"]), dim=1).to(torch.device("cpu"))
labels = data["label"].to(torch.device("cpu"))
else:
bands = torch.cat((data["bands10"], data["bands20"],data["bands60"]), dim=1).to(torch.device("cuda"))
labels = data["label"].to(torch.device("cuda"))
optimizer.zero_grad()
logits = model(bands)
loss = customLoss(logits, labels)
loss.backward()
optimizer.step()
lossTracker.update(loss.item(), numSample)
generated_codes += list(logits)
label_train += list(labels)
name_train += list(data['patchName'])
train_writer.add_scalar("loss", lossTracker.avg, epoch)
print('Train loss: {:.6f}'.format(lossTracker.avg))
#s = torch.sum(model.FC.weight.data)
#print('FC Sum Weight data: ',s)
return (generated_codes,label_train,name_train)
And the model:
class ResNet50PairWise(nn.Module):
def __init__(self, bits = 16):
super().__init__()
resnet = models.resnet50(pretrained=False)
self.conv1 = nn.Conv2d(12, 64, kernel_size=(7, 7), stride=(2, 2), padding=(3, 3), bias=False)
self.encoder = nn.Sequential(
self.conv1,
resnet.bn1,
resnet.relu,
resnet.maxpool,
resnet.layer1,
resnet.layer2,
resnet.layer3,
resnet.layer4,
resnet.avgpool
)
self.FC = nn.Linear(2048, bits)
self.apply(weights_init_kaiming)
self.apply(fc_init_weights)
def forward(self, x):
x = self.encoder(x)
x = x.view(x.size(0), -1)
logits = self.FC(x)
sign = torch.sign(logits)
binary_out = torch.relu(sign)
return binary_out
Thanks in advance