Hi i wanna apply data augmentation like i have an ecg dataset which is imbalanced. my question is if i have 1000, 5000, 6000, 7000, 6050, 5890 images in separate classes do i need augmentation? i mean how do you tell if you need? or it is only needed for highly imbalanced case like say 161 images in 1 class and rest are above 1000? 2ndly Now i want to apply stretching along xaxis keeping y axis constant. and then cropping image of original size. how can i achieve this??? because i have to be cautious about clipping of R peak.
To determine if data augmentation is needed, you can train your model without augmentation and analyze its performance. If the performance is satisfactory, you might not need augmentation. However, if your model is overfitting or its performance is not satisfactory, you can consider using data augmentation to help improve its performance.
To apply stretching along the x-axis while keeping the y-axis constant and then cropping the image to the original size, you can use the PIL library in Python. Here’s an example of how you can do this:
from PIL import Image
def stretch_image(image, stretch_factor):
original_width, original_height = image.size
new_width = int(original_width * stretch_factor)
# Stretch the image
stretched_image = image.resize((new_width, original_height), Image.ANTIALIAS)
# Crop the image to the original size while keeping the center intact
left = (stretched_image.width - original_width) / 2
top = (stretched_image.height - original_height) / 2
right = (stretched_image.width + original_width) / 2
bottom = (stretched_image.height + original_height) / 2
cropped_image = stretched_image.crop((left, top, right, bottom))
return cropped_image
# Load your image using PIL
image = Image.open("your_image_path.jpg")
# Apply the stretching and cropping
stretch_factor = 1.2
stretched_and_cropped_image = stretch_image(image, stretch_factor)
# Save the result
stretched_and_cropped_image.save("your_output_image_path.jpg")
be cautious about clipping the R peak in ECG images. You can adjust the cropping process to ensure the R peak remains intact. When using this augmentation technique, you can apply it to the images in the underrepresented classes to help balance the dataset. Also, keep in mind that you can combine this augmentation with other augmentation techniques such as rotation, flipping, or adding noise, to further increase the diversity of your dataset.
alright i did exactly this but i wanted to crop only starting part like i want to cropped the streteched image to the dimensions of original image. so i used 0,0,width,height as argument to .crop
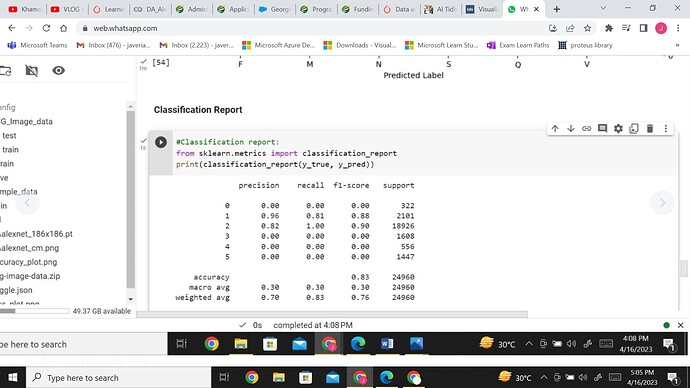
But now as i augmented my data from161 to 322 samples my classification report shows 0 precision and 0 ecall hence 0 f1score that is undefined. Have a look
You can see the support on right. Do you think this is due to imbalance? or as I m using alexnet as single channel grayscale 186x186 size so which is why???
To identify the root cause of the issue, you can try the following:
- Evaluate the performance of the model without augmentation and see if the issue persists. If the model performs better without augmentation, you might need to revise your data augmentation strategy.
- Experiment with different model architectures or fine-tune the existing model to better adapt it to your specific dataset.
- Use techniques like oversampling, undersampling, or other data augmentation methods to further balance the dataset and see if it improves the model’s performance.
By analyzing the results after each change, you can better understand the cause of the low precision, recall, and F1-score, and work towards improving your model’s performance.
Without having a look to the code it is hard for me to find the root cause… As you said you are using AlexNet, so alexnet was primarily designed for color images (with 3 channels), and as your dataset has grayscale images with a single channel. I am hoping you have modified the first layer of the model to accept a single channel input. Also, the size of the input images (186x186) is different from the default input size for AlexNet (224x224). I am hoping you have made these changes.
Yes I did the changes. Im not getting why this problem occured. When i did on mnist dataset it worked well but ecg data is highly imbalanced.