This is my main loop, it is a bit involved
for epoch in range(args.start_epoch, args.epochs):
t0 = time.time()
for i, (samples, target, idxs) in enumerate(train_loader):
t1 = time.time()
samples = samples.to(device, non_blocking=True)
target = target.to(device, non_blocking=True)
lambd_i = lambd[idxs, :]
lambd_i = lambd_i.to(device, non_blocking=True)
idxs = idxs.long()
adjust_learning_rate(optimizer, epoch, i, len(train_loader), args)
t2 = time.time()
uf_i = descriptor(samples)
u_i = classifier(uf_i)
t3 = time.time()
if use_admm_features:
vf_i = vf[idxs, :]
vf_i = vf_i.to(device, non_blocking=True)
v_i = classifier(vf_i)
else:
v_i = v[idxs, :].to(device, non_blocking=True)
labeled_batch_size = target.data.ne(NO_LABEL).sum().item()
batch_size = len(target)
misfit_k = misfit_fnc_k(u_i, target) / labeled_batch_size
misfit_u = rho * misfit_fnc_u(input=u_i, target=v_i, weight=cardinal_weights_u) / batch_size
lagrange = lagrange_fnc(lambd_i, u_i, v=v_i,rho=rho,weight=cardinal_weights_u) / batch_size
loss = misfit_k + misfit_u + lagrange
meters.update('misfit_k', misfit_k, labeled_batch_size)
meters.update('misfit_u', misfit_u, batch_size)
meters.update('lagrange', lagrange, batch_size)
meters.update('time_data', lagrange, batch_size)
t4 = time.time()
optimizer.zero_grad()
loss.backward()
optimizer.step()
t5 = time.time()
meters.update('time_data', t5-t1)
# print("loader = {:.2E}, rest = {:.2E}".format(t1-t0,t5-t1))
# t0 = time.time()
meters.update('time_tot',t5-t0)
LOG.info('misfit_k: {meters[misfit_k]:.4f} \t'
'misfit_u: {meters[misfit_u]:.4f} \t'
'lagrange: {meters[lagrange]:.4f} \t'
'time_data: {meters[time_data]:.4f} \t'
'time_tot: {meters[time_tot]:.4f} \t'.format(meters=meters))
meters.reset()
The dataset is litterally just scattered points lying in the 2D plane, so the features are x and y coordinates of each point, and the targets are then either 0,1,2 or -1 for noise.
Here is a visualization of my datapoints, each different class is a different color.
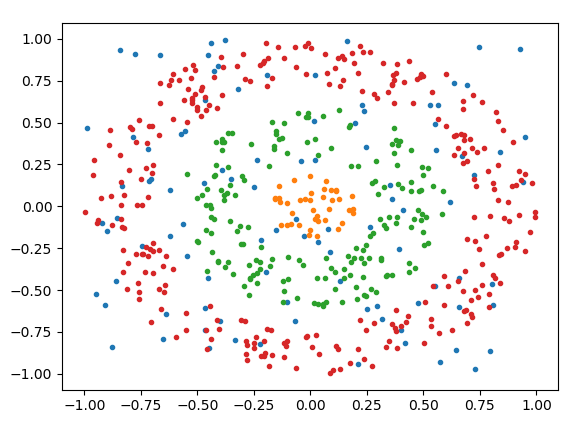
The database is created using this function:
class TensorDataset_custom(data.TensorDataset):
def __init__(self, samples,targets):
super(TensorDataset_custom, self).__init__(samples,targets)
self.targets = targets
self.samples = samples
classes = torch.unique(targets)
self.nclasses = sum(classes.numpy() >= 0)
def __getitem__(self, index):
sample = self.samples[index]
target = self.targets[index]
return sample, target, index
Regarding timing, I’m currently running everything on cpu in this simple example, so that shouldn’t be an issue

 I just did that small experiment and, with no preprocesing and reading from RAM, it’s not worth it to use
I just did that small experiment and, with no preprocesing and reading from RAM, it’s not worth it to use