I have been trying to fix this issue for some time and still have not managed. My main guess is that the problem resides in the DataLoader, as the class were I’m loading the data, prints the paths and pointclouds properly!
I am trying to train a network (PCN Pytorch) with 2 inputs: Partial PointClouds (Input) and their labels Complete PointClouds (Ground Truth). Something quite strange is happening: After the first epoch, the Complete PCs get ‘corrupted’. It is doubly strange as the partials are always different, but because of the design of what I am doing, the complete PC of an object is always the same! So I tried loading always 1 complete per object, N times (being N the amount of partials per object as well), and loading N completes (the same one), in case it was a problem of 'trying to open the same file concurrently).
train_dataset = ShapeNet('data/PCN', 'train', params.category)
train_dataloader = DataLoader(train_dataset, batch_size=params.batch_size, shuffle=True, num_workers=params.num_workers)
I have been checking the data up to Shapenet’s __getitem__(self, index), and visualizing those pointclouds, I can tell they have no errors. The dataset is relatively small so I can check them manually at the moment.
def __getitem__(self, index):
partial_path = self.partial_paths[index]
complete_path = self.complete_paths[index]
partial_pc = self.random_sample(self.read_point_cloud(partial_path), 2048)
complete_pc = self.random_sample(self.read_point_cloud(complete_path), 16384)
## To visualize the complete PC:
# pcd = o3d.geometry.PointCloud()
# pcd.points = o3d.utility.Vector3dVector(complete_pc)
# o3d.visualization.draw_geometries([pcd])
##
return torch.from_numpy(partial_pc), torch.from_numpy(complete_pc)
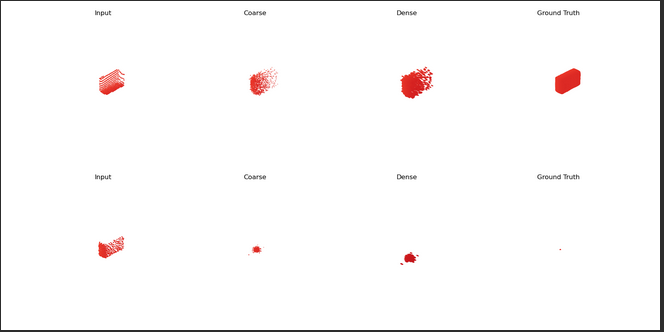
This is my main source of headaches. But I thought I could somewhat fix it. I realised that after using dataloader, when trying to visualise those Pointclouds in the for i, (p, c) in enumerate(train_dataloader): (p of partial, c of complete) in the training process:
for epoch in range(1, params.epochs + 1):
# hyperparameter alpha
if train_step < 10000:
alpha = 0.01
elif train_step < 20000:
alpha = 0.1
elif epoch < 50000:
alpha = 0.5
else:
alpha = 1.0
# training
model.train()
for i, (p, c) in enumerate(train_dataloader):
p, c = p.to(params.device), c.to(params.device)
optimizer.zero_grad()
# forward propagation
coarse_pred, dense_pred = model(p)
# loss function
After visualizing c during the first epoch, Complete PCs were alright. After the first epoch, they were as shown in the picture, regardless the object. I thought: well, I might be able to load it (as it’s just one per object) in here, convert it to a tensor, and substituting c in each iteration, but I am also not managing.
complete_pc = random_sample(read_point_cloud(path), 16384)
c = torch.from_numpy(complete_pc)
c = c.to(params.device)
Why is DataLoader doing this? Why would it load correctly all partials, but on the other side, the complete PCs would work only in the first epoch?