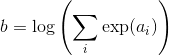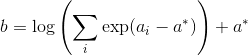I am trying to train a Mixture Density Network, by reproducing the results of the toy example of section 5 of Bishop’s paper, where MDNs were initially proposed (link to the paper: https://publications.aston.ac.uk/373/1/NCRG_94_004.pdf).
I am using exactly the same network architecture proposed there (single hidden layer with 20 neurons and tanh activation). The only difference is that I am using SGD instead of BFGS as my optimization algorithm.
Unfortunately, after 2k or 3k iterations (where the loss reduces considerably), I start getting NaN’s as the loss value. After some intense debug, I finally found out where these NaN’s initially appear: they appear due to a 0/0 in the computation of the gradient of the loss w.r.t. the means of the gaussian.
I am using negative log-likelihood as the loss function, L=-sum(log(p_i)). Therefore, the gradient of L w.r.t. p_i is
dLdp= -1/p_i
However, the derivative of a Gaussian function w.r.t. its mean, mu, is:
dpdmu = p_i * (x_i-mu_i)/(sigma_i^2)
so it is proportional to p_i. Therefore, when p_i is close to 0, the derivative of the loss w.r.t. the mean is:
dLdmu = dLdp * dpdmu = 0/0 -> NaN
However, this indetermination is easy to eliminate, since the expression may be algebraically simplified to dLdmu = (mu_i - x_i)/sigma_i^2. Of course, all that PyTorch does is numeric computing, so it is not able to do this simplification. How can I deal with this issue? Can I at least replace NaN’s with something else (zeros, for instance), so that they do not propagate?
(In all my reasoning I have assumed a MDN with one single Gaussian kernel, which is kind of stupid, but similar results roughly apply if we consider multiple kernels.)
Thank you in advance.