I read through a related thread and didn’t quite get to an answer so I thought I’d try and raise this issue on my own:
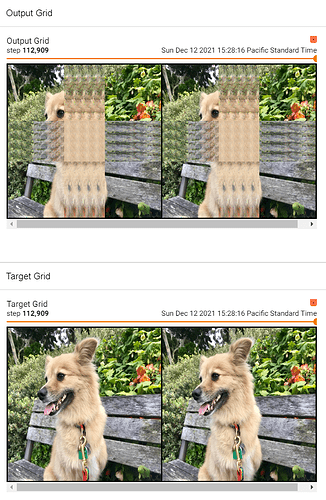
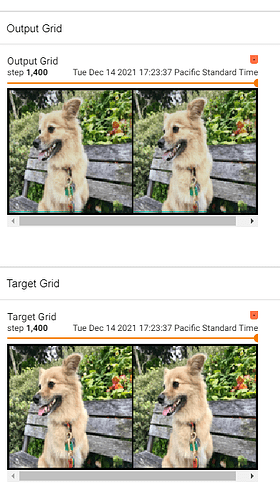
I tried implementing a UNet for image-to-image learning. To debug it I’m trying to overfit on a single example. It learns most of the image perfectly well, but the area where the convolutions seem to overlap in the final image are encountering problems. I’m not sure where my issue is.
One can see the top (output now) versus the bottom (as it should be).
I have a full implementation and the related training code uploaded here: GitHub - JosephCatrambone/UNet
But here is the relevant model architecture:
class DoubleConv(nn.Module):
def __init__(self, in_channels: int, out_channels: int):
super(DoubleConv, self).__init__()
self.op = nn.Sequential(
# No bias 'cause we're using BatchNorm. It will get cancelled out.
nn.Conv2d(in_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False),
nn.BatchNorm2d(out_channels),
nn.LeakyReLU(inplace=True),
nn.Conv2d(out_channels, out_channels, kernel_size=3, stride=1, padding=1, bias=False),
nn.BatchNorm2d(out_channels),
#nn.LeakyReLU(inplace=True),
)
def forward(self, x):
return self.op(x)
class UNet(nn.Module):
def __init__(self, in_channels:int = 3, out_channels: int = 3, feature_counts=None):
super(UNet, self).__init__()
# Prevent modification of mutable default.
if feature_counts is None:
feature_counts = [64, 128, 256, 512]
# Need nn.ModuleList instead of List for batch evals.
self.downsamples = nn.ModuleList()
self.bottleneck = DoubleConv(feature_counts[-1], feature_counts[-1]*2)
self.upsamples = nn.ModuleList()
self.finalconv = nn.Conv2d(feature_counts[0], out_channels, kernel_size=1) # 1x1 conv -> Change # feats.
self.pool = nn.MaxPool2d(kernel_size=2, stride=2)
# Downsample-Reduction step.
num_channels = in_channels
for f_count in feature_counts:
self.downsamples.append(DoubleConv(in_channels=num_channels, out_channels=f_count))
num_channels = f_count
# Up-steps.
for f_count in reversed(feature_counts):
self.upsamples.append(nn.ConvTranspose2d(f_count*2, f_count, kernel_size=2, stride=2))
self.upsamples.append(DoubleConv(f_count*2, f_count))
def forward(self, x):
skip_connections = list() # Don't need ModuleList because this is not retained.
for dwn in self.downsamples:
x = dwn(x)
skip_connections.append(x)
x = self.pool(x)
x = self.bottleneck(x)
skip_connections.reverse()
for idx in range(0, len(self.upsamples), 2):
x = self.upsamples[idx](x)
sk = skip_connections[idx//2]
# It's possible that due to integer division the sizes slightly mismatch.
if x.shape != sk.shape:
x = torch.nn.functional.interpolate(x, size=sk.shape[2:])
assert len(sk.shape) == 4 # So we don't accidentally unpinch another dimension.
concat_skip = torch.cat((sk, x), dim=1) # Dim 1 is channel-dimension. [b, c, h, w]
x = self.upsamples[idx+1](concat_skip)
return self.finalconv(x)
My best guesses for the source of the issue are something to do with the upscaling and the concatenation – maybe I’m joining along the wrong axis. Or, more likely, there’s something wrong with the padded convolution, but I can’t seem to make it work without the same-size conv2d.
Any input or debugging tips would be appreciated.