Hi,
Overview
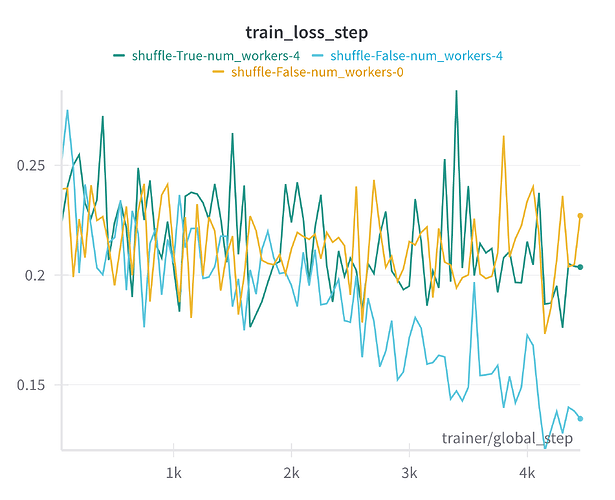
Recently I have encountered some strange overfitting when setting num_workers > 0. It turned out we accidentally had shuffle=False, but our models were still training fine when num_workers=0. What’s strange is when we set num_workers > 0, the models started to overfit – training loss was going down, but validation loss started to go up after a while. We solved it by setting shuffle=True; but I would still be interested in getting some insight into why this happened, because I find it strange that no shuffling worked fine, but then possibly a little bit of shuffling or data duplication introduced by the distributed training caused overfitting, but then fully shuffling the data caused it to train normally again.
Some background
We are training an algorithm called Noise2Void, which is a self-supervised method for denoising images that have uncorrelated noise. The basic idea is that it cannot learn the uncorrelated noise, but it can learn underlying structures present in an image. This means if we train using L2 loss it should learn to predict the expected mean of a pixel given the structure contained in some surrounding pixels.
This is implemented as part of our Python library CAREamics, that aims to make this algorithm (N2V) and others more accessible to the scientific community.
Example result using DataLoader parameters num_workers=4 and shuffle=False
(Bad result)
Side note
We are using PyTorch Lightning and we accidentally didn’t have shuffle=True because of a misunderstanding that PyTorch Lightning automatically applies shuffling to the train dataloader by using a torch.utils.data.DistributedSampler. After some further investigation it turned out that the data is in fact not shuffled during training unless shuffle=True is explicitly set in the train_dataloader.
I haven’t brought this up in the Lightning forums because I don’t believe it is a result of lightning, but if it turns out it is, I will bring it up there.
More notes
This is also raised as an issue on our GitHub repo where you can see good training results examples, and also figures plotting the train loss and validation loss.
Thanks!