Hi @ptrblck,
Thank you so much for your assistance!
I followed your recommendation and found that the model was busted and only outputting 0’s. After some review it seems that I fundamentally misunderstood the difference between creating a binary classification and multi-class classification model. Specifically, I was trying to use “One Hot Encoded” data with a softmax activation layer, and BCEwithlogits loss. I was able to run the model after setting the model to one class while using a sigmoid activation layer and BCE loss.
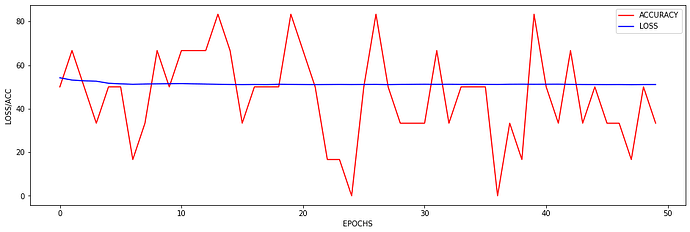
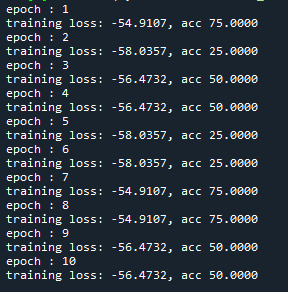
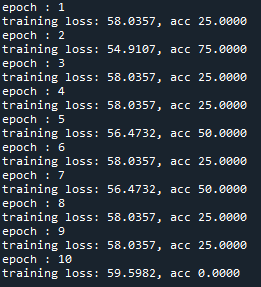
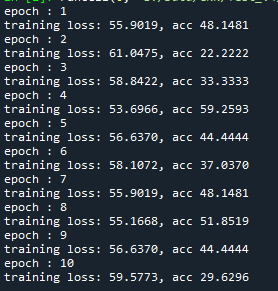
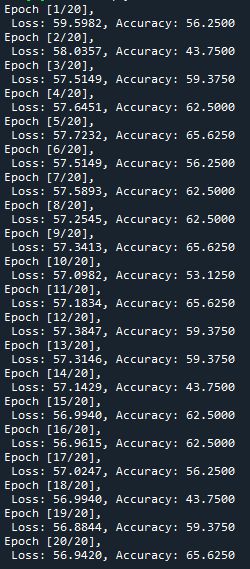
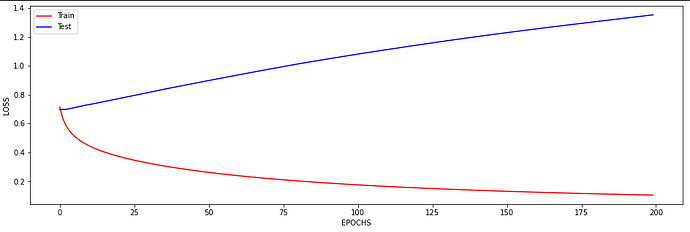
At the moment the training looks as follows:
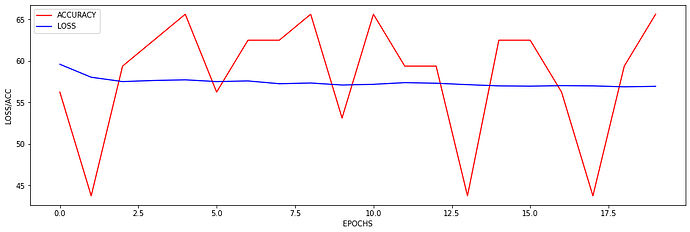
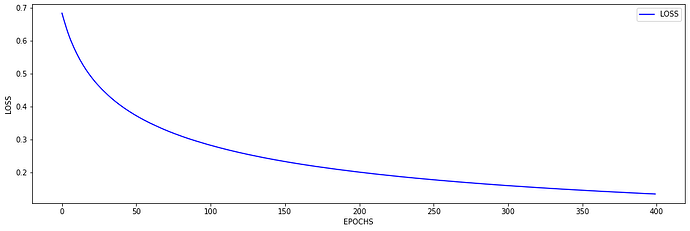
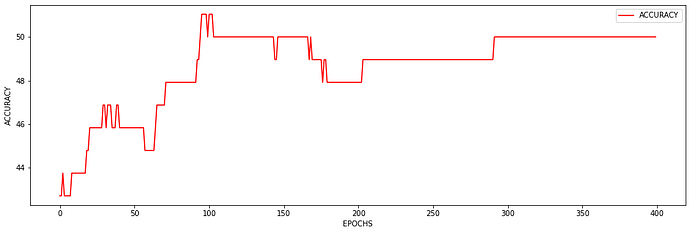
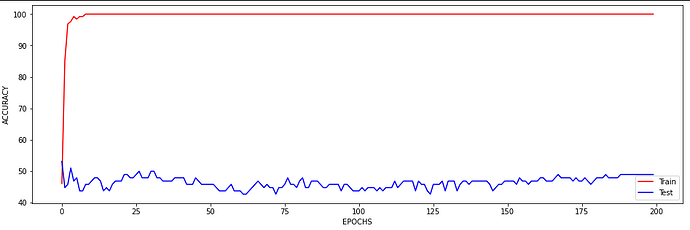
I just wanted to inquire about the accuracy. It is very blocky and gives poor results (chance-like accuracy). I have had success getting the loss to smoothly decline; however, I haven’t been able to get the accuracy to gradually and smoothly increase as I’ve seen in other’s results. Do you have any suggestions for how to approach this problem? Or is it more a matter of tweaking hyper-parameters?
At the moment I find that I can stop the accuracy from being so jumpy by increasing the batch size to 128 (see accuracy figure above) and it gradually increases with a learning rate of 0.001. If I change the learning rate the accuracy either decreases (0.01) or remains constant (0.0001). maybe I need a more complex model, momentum, or even data augmentation (I currently have 480 samples)? I am unsure.
I will reattach my code for my model and below for those observing this post so they can see the changes I’ve made.
Dataloader: Dataloader_V2 - Pastebin.com
Model:
import torch
from torch import nn
import torchmetrics
class CNN_model(nn.Module):
def __init__(self):
super(CNN_model, self).__init__()
self.convolutional_layer = nn.Sequential(
nn.Conv3d(in_channels=8, out_channels=16, kernel_size=(8, 8, 8), dilation=(3, 3, 3), stride=(2, 2, 2),
padding=(0, 0, 0)),
nn.BatchNorm3d(16),
nn.ReLU(),
nn.Conv3d(in_channels=16, out_channels=24, kernel_size=(8, 8, 8), dilation=(2, 2, 2), stride=(2, 2, 2),
padding=(0, 0, 0)),
nn.BatchNorm3d(24),
nn.ReLU(),
nn.Conv3d(in_channels=24, out_channels=32, kernel_size=(3, 3, 6), dilation=(1, 1, 1), stride=(1, 1, 1),
padding=(0, 0, 0)),
nn.BatchNorm3d(32),
nn.ReLU()
)
self.linear_layer = nn.Sequential(
nn.Linear(in_features=32, out_features=24),
nn.BatchNorm1d(24),
nn.ReLU(),
nn.Linear(in_features=24, out_features=16),
nn.BatchNorm1d(16),
nn.ReLU(),
nn.Linear(in_features=16, out_features=1))
#self.accuracy = torchmetrics.Accuracy()
def forward(self, x):
x = self.convolutional_layer(x)
x = torch.flatten(x, 1)
x = self.linear_layer(x)
x = torch.sigmoid(x) # must have dim =1 for softmax https://discuss.pytorch.org/t/bceloss-vs-bcewithlogitsloss/33586
return x
Training/Testing file:
import torch
import torchvision.transforms as transforms
import numpy as np
from torch.autograd import Variable
from numpy import vstack
from sklearn.metrics import accuracy_score
from matplotlib import pyplot as plt
from torch.utils.data.sampler import SubsetRandomSampler
from dataloader import RandomFmriDataset
from model import CNN_model
"""
Use "torch.cuda.empty_cache()" if you have the CUDA out of memory RuntimeError
"""
if __name__ == '__main__':
#Defining hyperparameters
learning_rate = 1e-3 #1e-2 results in decreseaing accuracy and 1e-4 in constant accuracy.
batch_size = 128 #higher batch size makes accuracy the same or smoothes it out. https://medium.com/mini-distill/effect-of-batch-size-on-training-dynamics-21c14f7a716e
num_epochs = 200
validation_split = .2
shuffle_dataset = True
random_seed= 42
num_classes = 1
compose = transforms.Compose([
transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))
]) #took out transforms.RandomHorizontalFlip(),
#Defining key dependencies
dataset = RandomFmriDataset()
device = torch.device("cuda")
model = CNN_model().cuda()
#Creating data indices for training and validation splits:
dataset_size = len(dataset)
indices = list(range(dataset_size))
split = int(np.floor(validation_split * dataset_size))
if shuffle_dataset :
np.random.seed(random_seed)
np.random.shuffle(indices)
train_indices, val_indices = indices[split:], indices[:split]
#Creating the data samplers and loaders:
train_sampler = SubsetRandomSampler(train_indices)
validation_sampler = SubsetRandomSampler(val_indices)
train_loader = torch.utils.data.DataLoader(dataset, batch_size=batch_size,
sampler=train_sampler)
validation_loader = torch.utils.data.DataLoader(dataset, batch_size=batch_size,
sampler=validation_sampler)
#Model training
def train_model(train_dl, model):
criterion = torch.nn.BCELoss(reduction='mean').to(device)
optimizer = torch.optim.SGD(model.parameters(), lr=learning_rate)
loss_array = []
loss_values = []
acc_values = []
for epoch in range(num_epochs):
for ind, (data, img_label) in enumerate(train_dl):
inputs = data.permute(0, 1, 4, 2, 3),
inputsV, labelsV = Variable(inputs[0]), Variable(img_label)
inputsV = inputsV.float()
labelsV = labelsV.float()
inputsV = inputsV.to(device)
labelsV = labelsV.to(device)
y_pred = model(inputsV)
loss = criterion(y_pred, labelsV)
running_loss = abs(loss.item())
loss_array.append(running_loss)
optimizer.zero_grad()
loss.backward()
optimizer.step()
avg_loss = np.average(loss_array)
loss_values.append(avg_loss)
acc = evaluate_model(validation_loader, model)
acc = acc * 100
acc_values.append(acc)
print(f'Epoch [{epoch + 1}/{num_epochs}], \n Loss: {abs(avg_loss):.4f}, Accuracy: {acc:.4f}')
acc_plot(acc_values)
loss_plot(loss_values)
#Model testing
def evaluate_model(test_dl, model):
predictions, actuals = list(), list()
accuracy_array = []
for ind, (data, img_label) in enumerate(test_dl):
inputs = data.permute(0, 1, 4, 2, 3),
inputsV, labelsV = Variable(inputs[0]), Variable(img_label)
inputsV = inputsV.float()
labelsV = labelsV.float()
inputsV = inputsV.to(device)
labelsV = labelsV.to(device)
yhat = model(inputsV)
actual = labelsV.cpu().numpy()
actual = actual.reshape((len(actual), 1))
yhat = yhat.cpu()
yhat = yhat.detach().numpy()
yhat = yhat.round()
predictions.append(yhat)
actuals.append(actual)
predictions, actuals = vstack(predictions), vstack(actuals)
acc = accuracy_score(actuals, predictions)
accuracy_array.append(acc)
#troubleshooting print commands for seeing model predictions and labels
'''
print(predictions)
print(labelsV)
'''
print(acc)
return acc
#Accuracy plot over epochs
def acc_plot(x):
plt.figure(figsize=(16, 5))
plt.xlabel('EPOCHS')
plt.ylabel('ACCURACY')
plt.plot(x, 'r', label='ACCURACY')
plt.legend()
plt.show()
#Loss plot over epochs
def loss_plot(x):
plt.figure(figsize=(16, 5))
plt.xlabel('EPOCHS')
plt.ylabel('LOSS')
plt.plot(x, 'b', label='LOSS')
plt.legend()
plt.show()
#The command for training the model
train_model(train_loader, model)
#The command for testing the model
evaluate_model(validation_loader, model)