I’m trying to generate one model’s parameters (ActualModel) with another model (ParameterModel), but running into problems with autograd when I backpropagate multiple times.
Here’s an example ActualModel, but this is supposed to be generic:
class ActualModel(torch.nn.Module):
def __init__(self):
super().__init__()
self.conv = torch.nn.Conv2d(1, 1, 3)
def forward(self, x):
return self.conv(x)
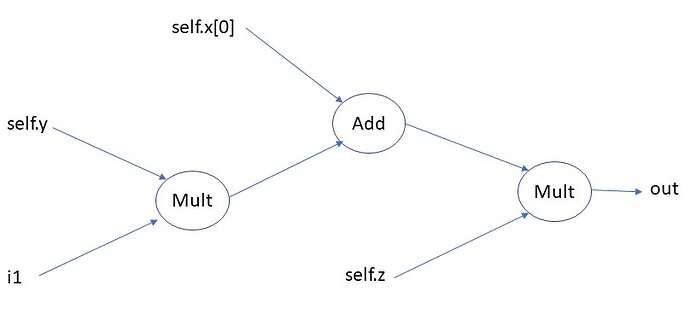
The ParameterModel wraps the ActualModel, freezes its parameters and in the forward() method, first computes the parameters, sets the values inside ActualModel and then computes ActualModel.forward():
class ParameterModel(torch.nn.Module):
def __init__(self, actual_model):
super().__init__()
# Save actual_model in a list so its parameters aren't auto-registered
self.actual_model = [actual_model]
# Compute total number of parameters of the actual_model
num_parameters = np.sum([p.numel() for p in actual_model.parameters()])
# Freeze parameters of actual_model
for p in actual_model.parameters():
p.requires_grad_(False)
# Simple model to generate num_parameters parameters
self.linear = torch.nn.Linear(1, num_parameters)
def forward(self, param_input, actual_input):
# Compute the parameters for the actual model
param_output = self.linear(param_input)
# Set the parameters in the actual model
idx = 0
for p in self.actual_model[0].parameters():
# p.set_(x) derivative is not implemented, use p *= 0, p += x instead
p.zero_().add_(param_output[idx:idx+p.numel()].view(p.shape))
idx += p.numel()
# Compute the output of the actual model
return self.actual_model[0](actual_input)
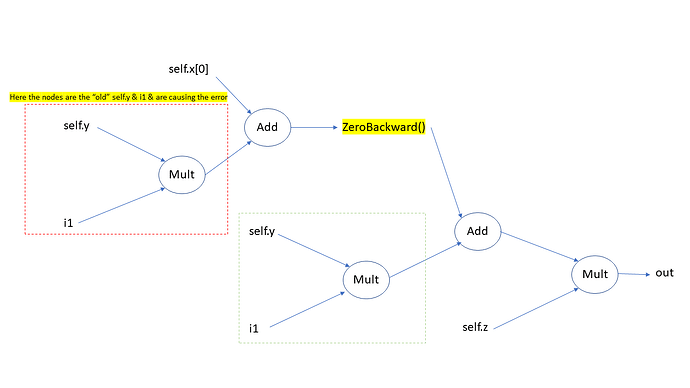
However, when I use this in a simplified training loop, on the second epoch I get an error about backwarding through the graph a second time, even though it should be a new graph in the second epoch.
Training loop:
actual_model = ActualModel()
wrapped_model = ParameterModel(actual_model)
optim = torch.optim.Adam(wrapped_model.parameters(), lr=0.07)
for i in range(2):
print(f'Epoch {i}')
wrapped_model.zero_grad()
output = wrapped_model(torch.ones(1), torch.ones(1, 1, 3, 3))
loss = output - torch.ones(1, 1, 3, 3)
loss.sum().backward()
optim.step()
Output with error message:
Epoch 0
Epoch 1
RuntimeError: Trying to backward through the graph a second time,
but the saved intermediate results have already been freed. Specify
retain_graph=True when calling backward the first time.
Any help on what I’m doing wrong here would be much appreciated.