import random
import torch
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
from torch_geometric.loader import DataLoader
from torch_geometric.data import Data
from torch_geometric.nn import GINConv, VGAE
from torch_geometric.nn.models import InnerProductDecoder
import matplotlib.pyplot as plt
device = torch.device(‘cuda:0’ if torch.cuda.is_available() else ‘cpu’)
batch_size = 32
class Encoder(nn.Module):
def __init__(self):
super(Encoder, self).__init__()
dim1 = 30
dim2 = 25
dim3 = 20
dim4 = 15
dim5 = 10
self.mlp1 = nn.Sequential(nn.Linear(dim1, dim2), nn.ReLU(),\
nn.Linear(dim2, dim3))
self.mlp2 = nn.Sequential(nn.Linear(dim1, dim2), nn.ReLU(),\
nn.Linear(dim2, dim3))
self.mlp3 = nn.Sequential(nn.Linear(dim3, dim5), nn.ReLU(),\
nn.Linear(dim5, dim5))
self.conv1 = GINConv(self.mlp1)
self.conv2 = GINConv(self.mlp2)
self.conv3 = GINConv(self.mlp3)
self.training = True
def reparametrize(self, mu, log_var):
if self.training:
std = torch.exp(0.5*log_var)
eps = torch.randn_like(std)
sample = mu+(eps*std)
else:
sample = mu
return sample
def forward(self, x, edge_index ):
# print('edge_index is', edge_index)
x = x.float()
mu = self.conv1(x,edge_index)
log_var = torch.tanh(self.conv2(x,edge_index)
lat_fea = self.reparametrize(mu, log_var)
return lat_fea, mu, log_var
class AdjacencyMatrix(nn.Module):
def forward_all(self, lat_fea, sigmoid = True):
'''
Parameters
----------
lat_fea
output from encoder
sigmoid : TYPE, optional
DESCRIPTION. The default is True.
Returns
-------
decodes the latent variables into a probablistic dense adjacency matrix
'''
adj_pred = torch.matmul(lat_fea, lat_fea.t())
pred_adjacency = torch.sigmoid(adj_pred)
return torch.sigmoid(adj_pred) if sigmoid else adj_pred
class VGAE(nn.Module):
def __init__(self):
super(VGAE, self).__init__()
self.encoder = Encoder()
self.adjacency = AdjacencyMatrix()
self.ns_loss = nn.BCELoss()
def forward(self, x, edge_index):
z = self.encoder.forward(x,edge_index)[0]
adj_pred = self.adjacency.forward_all(z)
return self.adjacency.forward_all(self.encoder(x, edge_index)[0])
def recons_loss(self, adj, adj_pred):
recon_loss = self.ns_loss(adj_pred, adj)
return self.ns_loss(adj_pred, adj)
def loss(self, ns_loss):
mu, log_var = self.encoder.forward(x, edge_index)[1], self.encoder.forward(x, edge_index)[2]
kld_new = -0.5 * torch.mean(torch.sum(1+2*log_var - mu**2 -log_var.exp()**2, dim = 1))
loss = ns_loss + kld_new
return ns_loss+ kld_new
batch_size = 32
states = torch.load(‘states.pt’).to(device)
x_o = states[:,:270].reshape(-1,9,30).to(device)
edge_index_o = states[:,391:431].reshape(len(x_o),2,-1).to(torch.long).to(device)
prev_job_o = states[:,270:390].reshape(-1,4,30).to(device)
cur_mach = states[:,390].reshape(len(x_o),1).to(device)
active_edges_o = states[:,431].reshape(len(x_o),1).to(device)
dataset = []
for _x,_edg,_active_edges in
zip(x_o,edge_index_o,active_edges_o):
adj = torch.zeros(9,9).to(device)
edges = _edg[:,:int(_active_edges)]
for i in edges.t():
adj[i[0]][i[1]] = 1
dataset.append(Data(x =_x, edge_index = _edg[:,:int(_active_edges)],
adj = adj))
random.shuffle(dataset)
split = int(len(dataset)*0.7)
train_dataset = dataset[:split]
test_dataset = dataset[split:]
test_loader = DataLoader(test_dataset, batch_size = 32, drop_last = True,
shuffle = True)
model = VGAE().to(device)
model.train()
model_optim = optim.Adam(model.parameters(), lr = 8e-04)
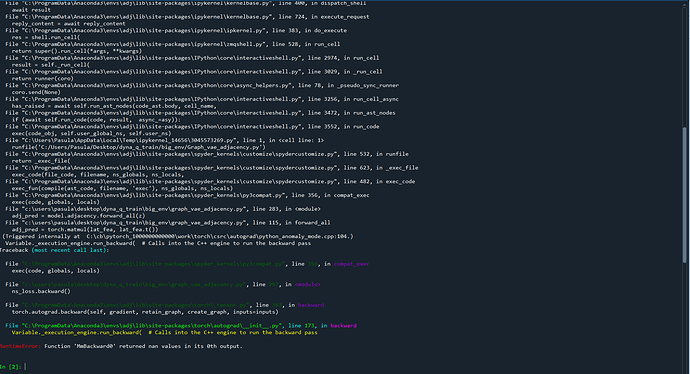
torch.autograd.set_detect_anomaly(True)
epoch = 1
epoch_counter = 0
tot_ns_loss = []
tot_acc = []
for ep in range(epoch):
train_loader = DataLoader(train_dataset, batch_size=1, drop_last = True,\
shuffle = True)
epoch_counter+=1
ep_acc = []
ep_ns_loss = 0
# ep_rew_loss = 0
count = 0
tot_loss = 0
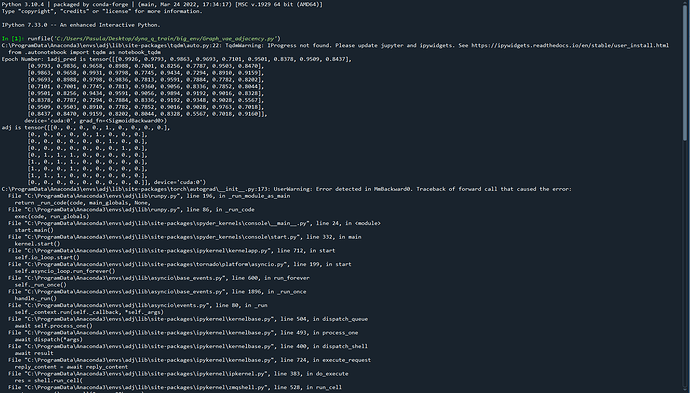
print('\rEpoch Number:', epoch_counter, end = "")
for data in train_loader:
count+=1
x, edge_index, adj = data.x, data.edge_index, data.adj
z = model.forward(x, edge_index)
adj_pred = model.adjacency.forward_all(z)
print('adj_pred is', adj_pred)
print('adj is', adj)
adj = adj.to(device)
ns_loss = model.recons_loss(adj_pred, adj)
ns_loss = model.loss(ns_loss)
ep_ns_loss += ns_loss.item()
model_optim.zero_grad()
ns_loss.backward()
model_optim.step()
tot_ns_loss.append(ep_ns_loss)
plt.title(‘ns_loss’)
plt.plot(tot_ns_loss)
plt.show()