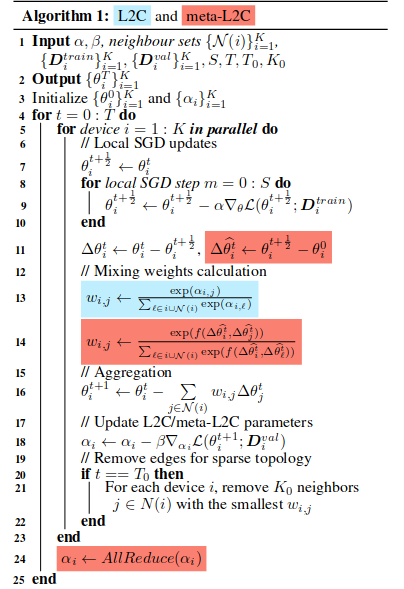
I am trying to implement this algorithm:
So I created a model like this:
class CNNCifar(nn.Module):
def __init__(self):
super(CNNCifar, self).__init__()
self.conv1 = nn.Conv2d(3, 6, 5)
self.pool = nn.MaxPool2d(2, 2)
self.conv2 = nn.Conv2d(6, 16, 5)
self.fc1 = nn.Linear(16 * 5 * 5, 120)
self.fc2 = nn.Linear(120, 84)
self.fc3 = nn.Linear(84, 10)
self.alpha = nn.Parameter(torch.randn(100, 100), requires_grad=True)
self.w = torch.randn((100, 100), requires_grad=True)
def forward(self, x):
x = self.pool(F.relu(self.conv1(x)))
x = self.pool(F.relu(self.conv2(x)))
x = x.view(-1, 16 * 5 * 5)
x = F.relu(self.fc1(x))
x = F.relu(self.fc2(x))
x = self.fc3(x)
return F.log_softmax(x, dim=1)
with a training loop that goes like this:
k = len(neighbour_sets)
device = torch.device("cuda" if not torch.cuda.is_available() else "cpu")
model = CNNCifar().to(device)
criterion = nn.CrossEntropyLoss()
optimizer = optim.SGD(model.parameters(), lr=0.1)
l2c_optimizer = optim.Adam([model.alpha], lr=beta, weight_decay=0.01)
test_accuracies = [[] for _ in range(k)]
theta = [model.state_dict().copy() for _ in range(k)]
theta_half = [model.state_dict().copy() for _ in range(k)]
# w = torch.randn(k, k, requires_grad=True)
delta_theta = [model.state_dict().copy() for _ in range(k)]
with tqdm_output(tqdm(range(T))) as trange:
for t in trange:
for i in range(k):
# Local SGD step
log.info(f'Started training a Local SGD at node {i + 1}')
model.load_state_dict(theta[i])
for m in range(S):
for _, data in enumerate(train_loaders[i]):
inputs, labels = data
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
outputs = model(inputs)
loss = criterion(outputs, labels)
loss.backward()
optimizer.step()
log.info(f'Finished training a Local SGD at node {i + 1}')
# Change capturing
log.info(f'Computing change capturing at node {i + 1}')
for name, param in model.named_parameters():
delta_theta[i][name] = theta[i][name] - theta_half[i][name]
log.info(f'Computing mixing weights at node {i + 1}')
# Mixing weights calculation
model.w = model.w.clone()
model.w[i] = compute_mixing_weights(model.alpha[i], neighbour_sets[i])
# Aggregation
log.info(f'Aggergating at node {i + 1}')
theta_next = {}
for name, param in model.named_parameters():
theta_next[name] = theta[i][name].clone()
for j in neighbour_sets[i]:
for name, param in model.named_parameters():
theta_next[name] -= model.w[i][j].item() * delta_theta[i][name][j].clone()
# Update L2C
log.info(f'Updating L2C at node {i + 1}')
model.load_state_dict(theta_next)
model.train()
# a training loop to find alpha that minimizes the validation loss
for _, data in enumerate(val_loaders[i]):
inputs, labels = data
inputs, labels = inputs.to(device), labels.to(device)
l2c_optimizer.zero_grad()
model.alpha.requires_grad_(True)
log.info(f'Forward pass check')
outputs = model(inputs)
loss = criterion(outputs, labels)
model.alpha.retain_grad()
loss.backward()
print(f'gradient of alpha is {model.alpha.grad}')
import pdb; pdb.set_trace()
l2c_optimizer.step()
# Update α[i]
# import pdb; pdb.set_trace()
# alpha_grad = model.alpha.grad # Access the computed gradients
# model.alpha.data[i] -= beta * alpha_grad[i]
# Remove edges for sparse topology
if t == T_0:
for _ in range(K_0):
j = min(neighbour_sets[i], key=lambda x: w[i][x])
neighbour_sets[i].delete(j)
theta[i] = model.state_dict().copy()
theta_half[i] = model.state_dict().copy()
# Compute test accuracy for each local model
test_accuracies = compute_test_acc(model, test_loaders[i], device, test_accuracies, i)
log.info(f'Test accuracies atiteration at Comm_round {t} = {sum(test_accuracies) / k}')
return theta, test_accuracies
The problem is: in step 18 of the algorithm, the gradient of the loss is computed with respect to alpha. but when I access the model.alpha.grad it’s None, an no gradient is available.
What am I doing wrong here?