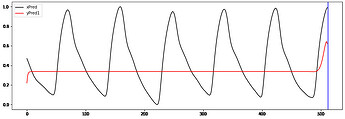
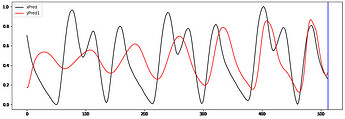
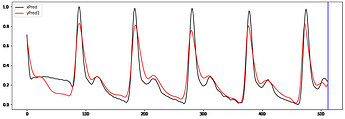
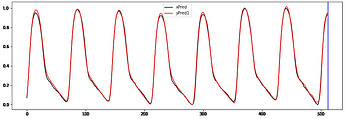
Hi to all,
Issue:
I’m trying to implement a working GRU Autoencoder (AE) for biosignal time series from Keras to PyTorch without succes.
The model has 2 layers of GRU.
The 1st is bidirectional.
The 2nd is not.
I take the ouput of the 2dn and repeat it “seq_len” times when is passed to the decoder.
The decoder ends with linear layer and relu activation ( samples are normalized [0-1])
I already try:
- Use the hidden of the 2d layer and pass it to the decoder and not the output.
- Quit and preserve the grad_clip functions
- Quit the 1st layer in decoder (gru_dec1)
I welcome any suggestions/advice
Model Keras:
inputs = Input(shape=(t, in_channels))
encoded = Bidirectional(GRU(256,return_sequences=True))(inputs)
encoded = GRU(32)(encoded)
decoded = RepeatVector(Signal_Len)(encoded)
decoded = GRU(32,return_sequences=True)(decoded)
decoded = Bidirectional(layers.GRU(256,return_sequences=True))(decoded)
decoded = TimeDistributed(layers.Dense(in_channels,activation=tf.nn.relu,
bias_initializer = b_init))(decoded)
Optimizer
opt = tf.keras.optimizers.RMSprop()
Loss
model_autoencoder.compile(optimizer=opt, loss=‘mse’, metrics=[‘mae’,‘mse’])
MODEL PYTORCH
Encoder
class EncoderRNN(nn.Module) def __init__(self, n_features, latent_dim, hidden_size): super(EncoderRNN, self).__init__() self.n_features = n_features self.hidden_size = hidden_size self.latent_dim = latent_dim self.gru_enc = nn.GRU(n_features, hidden_size, batch_first = True,dropout=0, bidirectional=True) self.lat_layer = nn.GRU(hidden_size*2, latent_dim, batch_first = True, dropout=0, bidirectional = False) def forward(self, x): x, _ = self.gru_enc(x) x , h = self.lat_layer(x) return x[:,-1].unsqueeze(1)
Decoder
class EncoderRNN(nn.Module): def __init__(self, seq_len, n_features , latent_dim , hidden_size): super(DecoderRNN, self).__init__() self.seq_len = seq_len self.n_features = n_features self.latent_dim = latent_dim self.hidden_size = hidden_size self.gru_dec1 = nn.GRU(latent_dim, latent_dim, batch_first = True, dropout=0, bidirectional= False) self.gru_dec2 = nn.GRU(latent_dim, hidden_size, batch_first = True, dropout=0, bidirectional= True) self.output_layer = nn.Linear(self.hidden_size*2, n_features,bias=True) self.act = nn.ReLU() def forward(self, x): x = x.repeat(1,self.seq_len, 1) x, _ = self.gru_dec1(x) x, _ = self.gru_dec2(x) return self.act(self.output_layer(x))
class AERNN(nn.Module): def __init__(self, seq_len, n_features, latent_dim , hidden_size): super(AERNN, self).__init__() self.seq_len = seq_len self.encoder = EncoderRNN(n_features, latent_dim, hidden_size).to(device) self.decoder = DecoderRNN(seq_len, n_features, latent_dim, hidden_size).to(device) def forward(self, x): x = self.encoder(x) x = self.decoder(x) return x
model = AERNN(seq_len, n_features, latent_dim , hidden_size) model.apply(bias_init) model = model.to(device) optimizer = torch.optim.RMSprop(model.parameters(), lr=1e-3) loss_fn = nn.MSELoss().to(device)
TRAIN
for epoch in range(100): #Train model.train() train_loss = 0 for x,_ in train_dl: x = x.cuda() optimizer.zero_grad() recon = model(x) loss = loss_fn(recon[:,:,0], x[:,:,0]) train_loss += loss.item() loss.backward() torch.nn.utils.clip_grad_norm_(model.parameters(), clip_norm) torch.nn.utils.clip_grad_value_(model.parameters(), clip_value) optimizer.step()