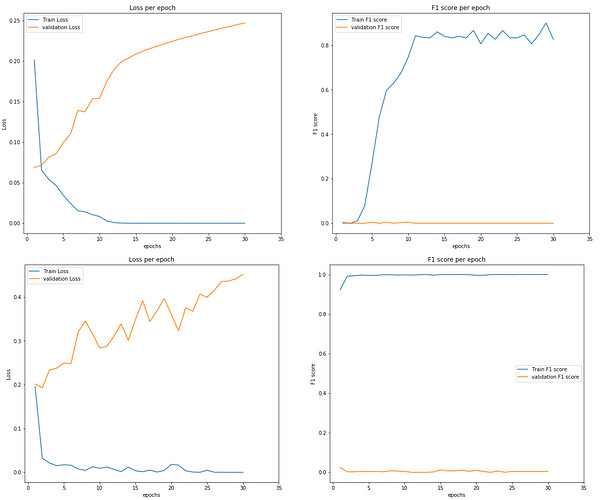
I’m working on a binary classification neural network with severe class imbalance (33130 zeros and 415 ones). While training my network, i test the accuracy for each epoch and it starts with 40%, reaches to 85% after 30 epochs. I’m using F1 score, recall and precision to measure the accuracy. the results vary a lot when i change the batch size of the dataloader.
when i feed the validation dataloader to the model however, accuracy plummets to 0%.
I don’t have the slightest idea what would be causing this, so ill try to include only essential information in this post while giving as much details as possible.
I started by splitting the dataset into features and labels, I then imputed the features, scaled it and reduced it using my SimpleImputer, StandardScaler and PCA objects
I then split the data using train_test_split
x_train, x_val, y_train, y_val = train_test_split(reduced_features,
labels,
test_size = 0.33,
random_state = 0)
I used WeightedRandomSampler to deal with class imbalance while following the guide in this article: PyTorch [Basics] — Sampling
target_list = torch.tensor(y_train.values)
target_list = target_list[torch.randperm(len(target_list))]
class_count = y_train.value_counts()
class_weights = 1./torch.tensor(class_count, dtype=torch.float)
class_weights_all = class_weights[target_list]
weighted_sampler = WeightedRandomSampler(
weights=class_weights_all,
num_samples=len(class_weights_all),
replacement=True
)
I created the following class to pass an object containing features and labels to the dataloader:
class Data_for_loader(Dataset):
def __init__(self, X_data, y_data):
self.X_data = X_data
self.y_data = y_data
def __getitem__(self, index):
return self.X_data[index], self.y_data[index]
def __len__ (self):
return len(self.X_data)
train_data = Data_for_loader(torch.FloatTensor(x_train.values),
torch.FloatTensor(y_train.values))
validation_data = Data_for_loader(torch.FloatTensor(x_val.values),
torch.FloatTensor(y_val.values))
I then created the dataloader objects:
train_loader = DataLoader(dataset = train_data, shuffle=False, batch_size = 150, sampler=weighted_sampler)
validation_loader = DataLoader(dataset = validation_data, batch_size= 1)
since this is a binary classification problem I’m using BCEWithLogitsLoss as my criterion
I’m using Adam as my optimizer
I created the following class for my model:
class BinaryClassifier(nn.Module):
def __init__(self):
super(BinaryClassifier, self).__init__()
self.h1 = nn.Linear(140, 90)
self.h2 = nn.Linear(90, 50)
self.output = nn.Linear(50, 1)
self.relu = nn.ReLU()
self.dropout = nn.Dropout(p=0.2)
self.batchnorm1 = nn.BatchNorm1d(90)
self.batchnorm2 = nn.BatchNorm1d(50)
def forward(self, x):
x = self.h1(x)
x = self.relu(x)
x = self.batchnorm1(x)
x = self.h2(x)
x = self.relu(x)
x = self.batchnorm2(x)
x = self.dropout(x)
x = self.output(x)
return x
I used the following code to train the model:
model.train()
for e in range(epochs):
epoch_loss = 0
recall_total_score = 0
precision_total_score = 0
f1_total_score = 0
for inputs, labels in train_loader:
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
output = model.forward(inputs)
loss = criterion(output, labels.unsqueeze(1))
#rounding output for accuracy measure
output = torch.round(torch.sigmoid(output))
output = output.cpu().detach().numpy()
labels = labels.cpu().unsqueeze(1)
#measuring accuracy
recall_total_score += recall_score(labels, output)
f1_total_score += f1_score(labels, output)
precision_total_score += precision_score(labels, output)
loss.backward()
optimizer.step()
epoch_loss += loss.item()
finaly i used the following code to evaluate the model:
y_pred = []
y_true = []
model.eval()
with torch.no_grad():
for inputs, labels in validation_loader:
inputs = inputs.to(device)
output = model.forward(inputs)
output = torch.round(torch.sigmoid(output))
y_pred.extend(output.cpu().numpy())
y_true.extend(labels.cpu().numpy())
f1 = f1_score(y_true, y_pred)
I’m relatively new to Pytorch, my only guess is that the model is overfitting, even tho I’m using dropout in my model, or i’m somehow making a mistake while dealing with the validation set.
Thanks for your help!