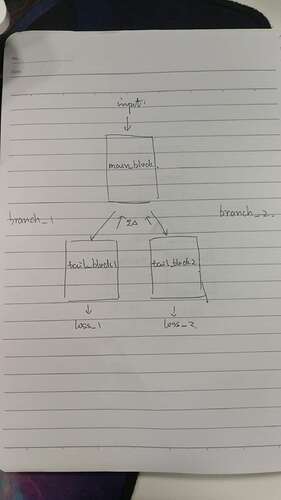
I am facing a multi-label problem. My network has 2 branches, which are separated from a conv layers.
There is the sketch of my network
class MultiLabelDemo(nn.Module):
def __init__(self):
super(MultiLabelDemo, self).__init__()
self.main_block = nn.Sequential(
nn.Conv2d(in_channels=3, out_channels=96, kernel_size=11, stride=4),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=96, out_channels=96, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=96, out_channels=96, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=3, stride=2, ceil_mode=True)
)
self.tail_block1 = nn.Sequential(
nn.Conv2d(in_channels=96, out_channels=256, kernel_size=5, stride=1, padding=2),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=3, stride=2, ceil_mode=True)
)
self.tail_block2 = nn.Sequential(
nn.Conv2d(in_channels=96, out_channels=256, kernel_size=5, stride=1, padding=2),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.Conv2d(in_channels=256, out_channels=256, kernel_size=1, stride=1),
nn.ReLU(inplace=True),
nn.MaxPool2d(kernel_size=3, stride=2, ceil_mode=True)
)
def forward(self, data):
x = self.main_block(data)
y1 = self.tail_block1(x)
y2 = self.tail_block2(x)
return y1, y2
branch_1 is for 1st label and branch is for 2nd label, so loss_1 is computed as
nn.CrossEntropyLoss(y1, label_1) and loss_2 is computed as nn.CrossEntropyLoss(y2, label_2).The whole network has only one optimizer (SGD).(I’m not sure whether it is correct.).
What I want to implement is that loss_1 is used to update the weights and bias in tail_block1 and loss_2 is used to update the weights and bias in tail_block2. when backprop progress goes to the layer when network was separated, add two grad together and update the remaining main_block. How can I do this, I’ve searched a lot but nothing found. I’d appreciate your kindly help.