i am working on miccai 2008 coronary artery tracking challenge dataset and I want to load it into pytorch then preprocessing it, but it is so complicated,
it is diveded as flowing :
training / dataset00 (from 00 to 08 ) / Dicom - seg (1 to 17 )
testing is the same
how to know the gorunthruth and validation for this data
How are the ground truth labels stored?
Do you have and file storing all labels or is the label encoded in the file names?
Most likely the best approach would be to create a custom Dataset, and load as well as preprocess your data in the __getitem__ method.
Have a look at the Data loading tutorial and let me know if you need some help implementing it.
the grountruth is stored as numbers in text doc
So you have one text document containing all ground truth labels?
I assume they are sorted somehow, i.e. the first number corresponds to the “first” input data?
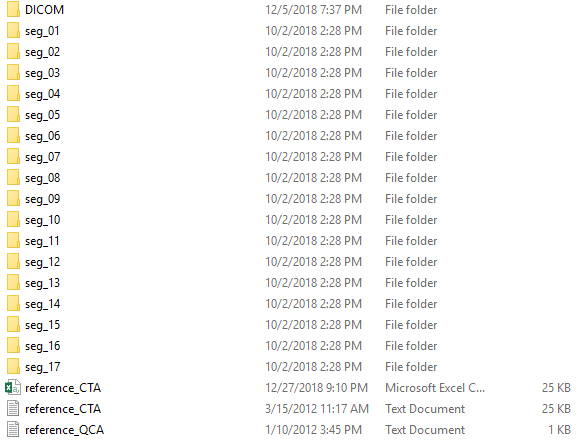
they are shown in the pic and there is ( 1 to 3 ) text doc in each ( seg_ ) file
Each of these text files in the seg_XX folders contains some target information for this segment, is that correct?
The seg_XX folders contain additionally some DICOM data or just the target?
Do you have any constraints regarding the datasetXX folders? E.g. would you like to handle them in a special way to avoid data leakage or can you just use all datasets to train your model?
the seg_xx folders contain just the target ( another refrence CTA ) .
all the datasets will be used to train the model
I can’t find much information on this particular dataset, so I would need some more input from you.
The seg_XX targets seem to have some kind of order. Are they just split to different slices in your DICOM data or do they have any temporal relationship?
What would be the basic workflow?
We would load the data stored in the DICOM folder, and load all seg_XX targets to create somehow a “global” target for this data?
acually i do not understand this dataset at all , but i was thinking of using reference.CAT file
in dataset_xx folder as grountruth for DICOM files in each dataset_xx folder
this is what is written in their website
" For each dataset, each directory segXX contains a file reference_CTA.txt that contains the CTA stenoses detection and quantification reference standard. The files contain the world position of the x-, y-, and z-coordinate of each path point of the segment centerline"
So you have a reference_CTA.txt in the datasetXX folder and additionally in each seg_XX folder?
We could load all of them or just the one in the top folder to create the ground truth.
Unfortunately, I don’t know, how this data should be handled.
yes there is reference_CTA.txt in the datasetXX folder and additionally in each seg_XX folder.
if we just load just the one in the top of the folder , should i combine them all in on file to make one ground truth?
This might work, but as I said I’m completely unfamiliar with this dataset.
Let me know, if you get stuck implementing some approach.
thank you @ptrblck , i am trying to implement your code
class MyDataset(Dataset):
def __init__(self, image_paths, target_paths, train=True):
self.image_paths = 'content/drive/My Drive/data/training/images'
self.target_paths = 'content/drive/My Drive/data/training/1st_manual'
def transform(self, image, mask):
# Resize
resize = transforms.Resize(size=(520, 520))
image = resize(image)
mask = resize(mask)
# Random crop
i, j, h, w = transforms.RandomCrop.get_params(
image, output_size=(512, 512))
image = TF.crop(image, i, j, h, w)
mask = TF.crop(mask, i, j, h, w)
# Random horizontal flipping
if random.random() > 0.5:
image = TF.hflip(image)
mask = TF.hflip(mask)
# Random vertical flipping
if random.random() > 0.5:
image = TF.vflip(image)
mask = TF.vflip(mask)
# Transform to tensor
image = TF.to_tensor(image)
mask = TF.to_tensor(mask)
return image, mask
def __getitem__(self, index):
image = Image.open(self.image_paths[index])
mask = Image.open(self.target_paths[index])
x, y = self.transform(image, mask)
return x, y
def __len__(self):
return len(self.image_paths)
but i am having this errro
def transform(self, image, mask):
^
IndentationError: unindent does not match any outer indentation level
also i want to display some of the images (DICOM format) to see how the prep works , how i can do that ?
It looks like __init__ and the other functions do not have the same indentation. Could you try to add a whitespace to __init__ or remove one from the others?
If you can successfully load the DICOM images using PIL you could also display it using image.show().
However, if that’s not working, you could try other packages like pydicom.
Hi,i need this dataset recently,but i can’t find any download links related to this dataset.Could you tell me,pls?
Have you downloaded the data set?
I need the CTA08 dataset, but it can not download anymore. Can you share it with me? Thank you!
Hi there, I am working on a quite interesting medical project in which one element of many is to perform centerline extraction. Do you still have the CAT08 dataset? It is sort of a benchmark set in many articles in this topic area. It would be more than useful to have this set to compare the results with the publications related to the MICCAI 2008 Coronary Artery Tracking Challenge challenge. The authors don’t respond, probably due to many such requests about this old project.
Please email me if possible p.tomasinski@gmail.com
I need the CTA08 dataset, but it can not download anymore. Can you share it with me? Thank you!my email is lishang0103@126.com
I need the CTA08 dataset, but it can not download anymore. Can you share it with me? Thank you!my email is my2002724@163.com