From my understanding, jax does as many backward as there are outputs just like @richard proposed. But they have the vmap operator that allows then to do this more efficiently than a for loop in python (even though the theoretical complexity is the same).
Do you know how their vmap works? I’m curious if it’s similar to what NestedTensor will eventually be or if they just do some program transformations to accomplish it
From my understanding: implement a batched version of every function: https://jax.readthedocs.io/en/latest/notebooks/How_JAX_primitives_work.html#Batching
I guess this post is related
And you might like the following two repos
Unfortunately, it doesn’t support all types of networks (e.g. no batch norm), but convnets and all types of activations will work with just one backprop.
@albanD @richard how does the linked gist work? Previously, when I tried running backward() twice, the model wouldn’t learn, but switching to autograd.grad() fixed whatever problem existed. If I can use the linked gist and get backward() to work when run twice, then I might have a solution!
Hi,
The difference is that .backward() accumulate gradients in the .grad fields of the leafs. .grad() does not.
So you were most likely accumulating extra gradients when doing multiple call to .backward() if you were not calling .zero_grad() before the last one.
Hmm… I’m not sure this accords with what I see empirically. I’m using grad() followed by loss.backward() and this seems to change the training of the model compared with just running loss.backward(). If grad doesn’t accumulate gradients, then why does the outcome differ?
Maybe I don’t know what exactly you mean by leafs.
Sorry maybe that may not have be clear. The two different cases are:
opt.zero_grad()
loss = xxx(inputs) # Compute your loss
grads = xxx(loss) # Compute gradients wrt your loss
penalty = xxx(grads) # Compute the gradient penalty
final_loss = loss + penalty
final_loss.backward()
opt.step()
In the example above, you want to make your gradient step only for the gradients computed during final_loss.backward(). But if the computation of grads is done with .backward(create_graph=True), then you accumulate some extra gradients. You don’t do this if you compute grads with autograd.grad(create_graph=True).
So the two gradients when you step are different. That could explain your model training properly in one case but not the other
@albanD @richard , I have a question regarding our above conversation. Referring to richard’s code near the top, I’m trying to rewrite his loop using a larger batch size, but I’m receiving One of the differentiated Tensors appears to not have been used in the graph. Set allow_unused=True if this is the desired behavior. Can you help me understand why I’m seeing this error?
Here’s what I’m doing. Let x be the input to the graph with shape (batch size, input dimension) and let y be the output of the graph with shape (batch size, output dimension). I then select a subset of N random unit vectors. I stack x with itself and y with itself as follows:
x = torch.cat([x for _ in range(N)], dim=0)
and
y = torch.cat([y for _ in range(N)], dim=0)
x then has shape (N * batch size, input dim) and y has shape (N * batch size, output dim). But then, when I try to use autograd, I receive the aforementioned error .
jacobian = torch.autograd.grad(
outputs=y,
inputs=y,
grad_outputs=subset_unit_vectors,
retain_graph=True,
only_inputs=True)[0]
RuntimeError: One of the differentiated Tensors appears to not have been used in the graph. Set allow_unused=True if this is the desired behavior.
Why is this, and is there a way to make this large batch approach work instead of looping?
It’s appears that repeating the tensor and concatenating the list destroys the path in the computational graph from x to y.
Are you sure it’s not a typo where you give y both to inputs and outputs in your call to autograd.grad() ?
I don’t think so. Here’s a minimal example:
import numpy as np
import torch
def get_batch_jacobian(net, x, to):
# to: total output dim
x.requires_grad_(True)
batch = x.shape[0]
y = net(x)
x_tiled = x.repeat(to, *(1,) * len(x.shape[1:]))
y_tiled = y.repeat(to, *(1,) * len(y.shape[1:]))
basis_vectors = torch.eye(to)
random_basis_indices = np.random.choice(
np.arange(to),
size=batch * to)
random_basis_vectors = basis_vectors[random_basis_indices]
test = torch.autograd.grad(
outputs=y_tiled,
inputs=x_tiled,
grad_outputs=random_basis_vectors,
retain_graph=True,
only_inputs=True)[0]
return test
class CNNNet(torch.nn.Module):
def __init__(self):
super(CNNNet, self).__init__()
self.cnn = torch.nn.Conv2d(1, 3, 5)
self.fc1 = torch.nn.Linear(3, 4)
def forward(self, x):
x = torch.nn.functional.relu(self.cnn(x))
x = x.reshape(x.shape[0], -1)
x = torch.nn.functional.relu(self.fc1(x))
return x
cnet = CNNNet()
batch = 10
x = torch.randn(batch, 1, 5, 5)
y = cnet(x)
ret = get_batch_jacobian(cnet, x, 4)
print(ret.shape)
The above code produces RuntimeError: One of the differentiated Tensors appears to not have been used in the graph. Set allow_unused=True if this is the desired behavior.
I suspect the reason is because PyTorch’s computational graph doesn’t understand the relationship between x and x_tiled. It probably thinks the graph looks like:
x -> y -> y_tiled
and
x -> x_tiled
So there’s no path from x_tiled to y_tiled, even though x_tilded has the same values that determined y_tiled.
Hi,
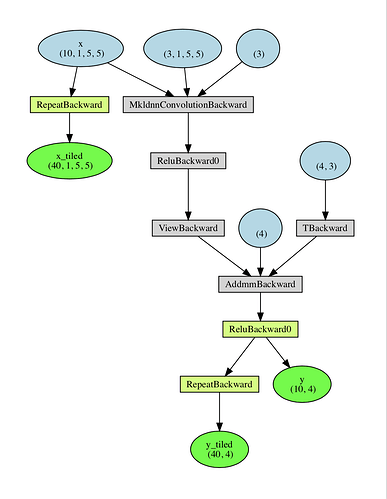
So the issue is that when you do .repeat(), you create a new differentiable function. And y does depend on x but not on x_tiled. See below the graph created by your code:
Let me know if you have more questions.
Ok this agrees with my hypothesis. Two questions:
-
how did you generate this graph?
-
is there any way to do what I’m trying to do?
Hi,
- https://github.com/szagoruyko/pytorchviz (slightly modified) and adding just before the autograd.grad:
import torchviz
params = {
"y_tiled": y_tiled,
"x_tiled": x_tiled,
"x": x,
"y": y
}
torchviz.make_dot((x_tiled, y_tiled, y), params=params).view()
- You will need to do the tiling of
xbefore you use it in the graph. So before you forward the net.
@albanD @richard quick question about Richard’s earlier post above in which he gave code to compute the Jacobian of a function that maps R^n to R^m. I need to compute the hidden state to hidden state Jacobian of a RNN with a single layer, evaluated at a number of points (i.e. over a batch), and I want to confirm that I’m doing this correctly.
The hidden state has shape (batch size, num layers=1, hidden state size). I first compute the unit basis vectors as
num_basis_vectors = num_layers * hidden_size
unit_basis_vectors = torch.eye(n=num_basis_vectors).reshape(
num_basis_vectors, num_layers, hidden_size)
I then loop over the unit basis vectors (as above):
jacobian_components = []
for unit_vector in unit_basis_vectors:
jacobian_component = torch.autograd.grad(
outputs=rnn_output_hidden_states, # shape (batch size, num layers=1, rnn hidden state size)
inputs=rnn_input_hidden_states, # shape (batch size, num layers=1, rnn hidden state size)
grad_outputs=torch.stack([unit_vector] * batch_size), # repeat batch_size times
retain_graph=True)[0]
jacobian_component = torch.mean(jacobian_component, dim=(1,))
jacobian_components.append(jacobian_component)
jacobian = torch.stack(jacobian_components, dim=1)
Is this correct? How would I go about testing the correctness?
Sorry about the bad indentation. I’m pasting from an IDE.