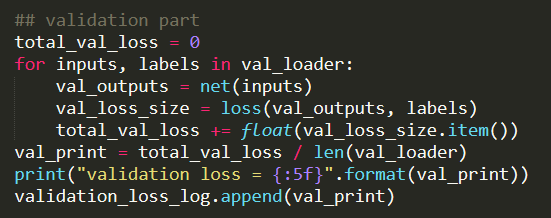
Hi ptrblck,
My implementation to create the dataloader may be naive and not clean to you.
1: To get data and label as numpy arrays: I use packages to read .dcm files and corresponding .nrrd files
2: I convert the numpy array (optionally crop or not) from defalut type unit16 to int16 in order to be used in pytorch:
below is one of the repetitive part for me to read .dcm file to get image array and convert a (320, 320) original image numpy array into (number of totoal image files,320,320):
''' > img_1 = [pydicom.dcmread(train_Prostate3T_img_path + '/' + ID + '/' + dcm_number).pixel_array for ID in Prostate3T_patient_ID for dcm_number in os.listdir(train_Prostate3T_img_path + '/' + ID )]
> Prostate3T_img = img_1[0][np.newaxis,...]
> count = len(Prostate3T_img)
> for i in range(1,len(img_1)):
> try:
> Prostate3T_img = np.vstack((Prostate3T_img, img_1[i][np.newaxis,...]))
> except:
> #print("mis-matched dimension at", i, "-th sample.")
> #print("wrong shape:", np.shape(img_1[i])) ## 18 wrong 256 x 256 shapes
> #print("prostate samples already counted:", count)
> continue
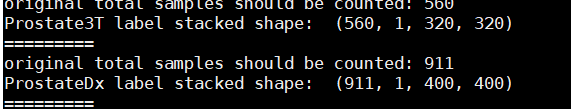
> print("original total samples should be counted:", np.shape(img_1)[0])
> print("Prostate3T image stacked shape: ", np.shape(Prostate3T_img))
> print("==="*3) '''
>
> Then I convert (1,320,320) per sample to (3,320,320) 3 channels using cv2.cvtColor:
> ''' img_to_3_channels_Prostate3T = np.array([cv2.cvtColor(Prostate3T_img[i], cv2.COLOR_GRAY2RGB).T for i in range(len(Prostate3T_img))])
''''
3: convert them into torch tensor and use .utils.TensorDataset to put in pytorch dataset
(the reason why I convert tensor type to .type(‘torch.FloatTensor’) is that I found I can only use L1 Loss in this way an gives no error, my custom loss function is still under deleloped)
img_to_3_channels_Prostate3T_int16 = np.array(img_to_3_channels_Prostate3T, dtype=np.int16)
img_Prostate3T_tensor = torch.from_numpy(img_to_3_channels_Prostate3T_int16)
img_Prostate3T_tensor = img_Prostate3T_tensor.type('torch.FloatTensor')
img_Prostate3T_tensor = img_Prostate3T_tensor.to(device)
Prostate3T_dataset = utils.TensorDataset(img_Prostate3T_tensor, label_Prostate3T_tensor)
- then use utils.DataLoader to establish a Dataloader with SubsetRandomSampler.
4.1 SubsetRandomSampler is used in this way:
train_sampler = SubsetRandomSampler(np.arange(n_training_samples, dtype=np.int64))
4.2. training set loader is used in this way:
train_loader = torch.utils.data.DataLoader(Prostate3T_dataset, batch_size=batch_size, sampler=train_sampler)
Any advice is appreciated!
Thank you very much!
Best,
Peter
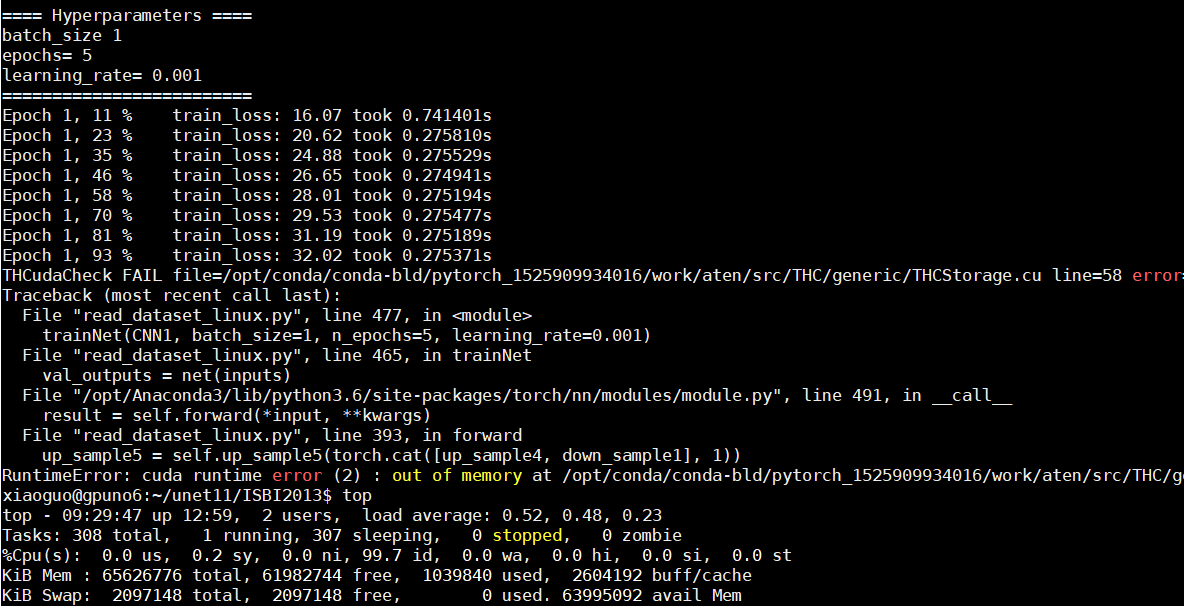

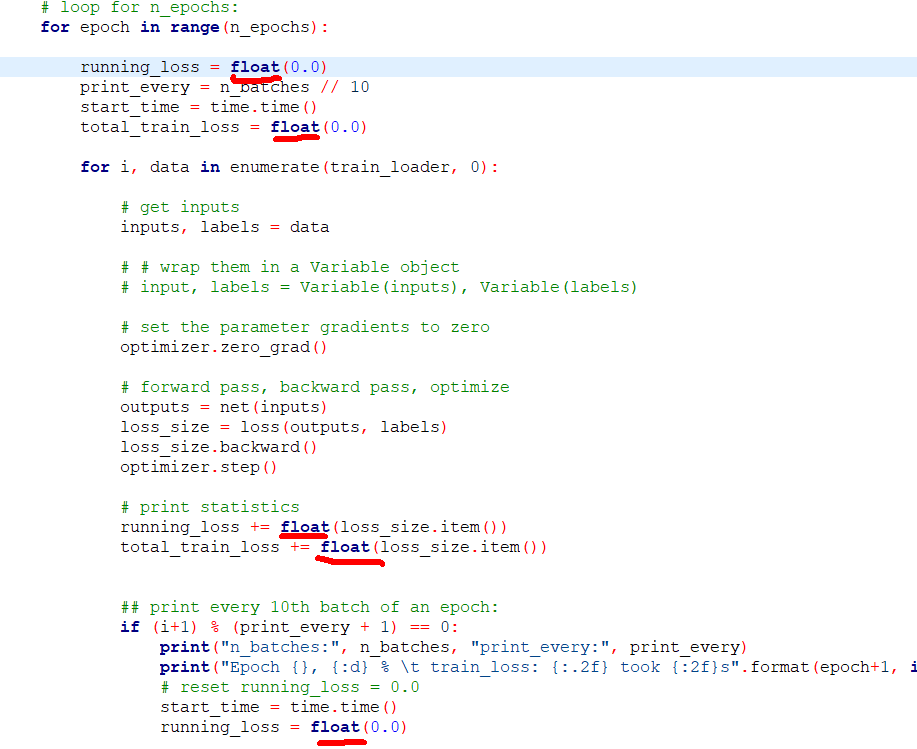
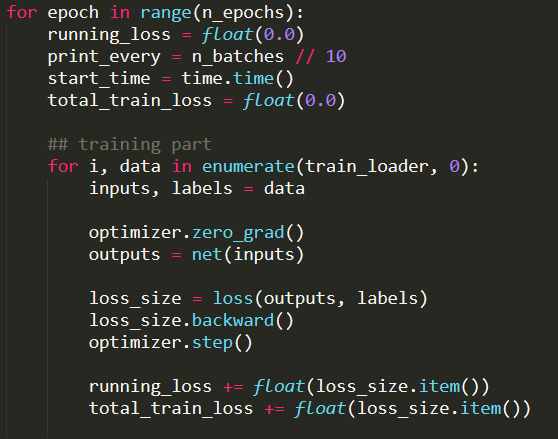
 , one last thing to look at is the train loader I think.
, one last thing to look at is the train loader I think.