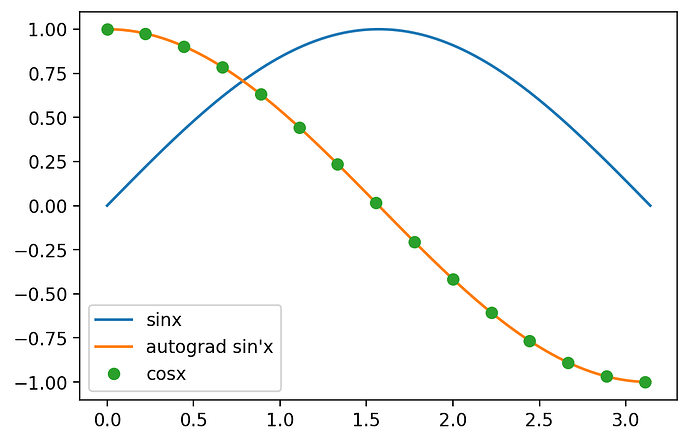
I would like to calculate y = sin(x), where x is a vector of length N and compute the corresponding derivative vector y’ = (dy_i / dx_i), i = 1,2,3,…,N using autograd.
This is what I ended up doing:
// Input vector x
auto x = torch::linspace(
0, M_PI, 100, torch::requires_grad()
);
// sin(x) as a vector-valued function
auto sinx = torch::sin(x);
// backpropagate so that the gradient value is stored at x
sinx.backward(torch::ones_like(x));
// x.grad() is dsin(x_i)/dx_i for some reason and not sinx.grad()
auto xgrad = x.grad();
// sin'(x) = cos(x) for comparison
auto cosx = torch::cos(x);
and it seems to work:
Is this correct and are there other ways (torch::autograd::grad) to calculate the gradient?
I have two questions.
-
Why do I have to give the argument
torch::ones_like(x)to thebackwardfunction ofsinx? I understand the backpropagation in a network, there’s the output given by the model, and the difference between the output and the target is proportional to the gradient at this layer with respect to the weights and biases, this is used to correct the network parameters, etc. Here there is simply x → sin(x),there aren’t any weights and biases in this “network” (AD graph). -
x → sin(x) is an acyclic directed graph, so why is the gradient of sin(x) stored at x? Why not at the sinx node? x.grad() somehow doesn’t intuitively mean sin’(x). Is x.grad() here the gradient of the root node (sinx) with respect to x, or did I misunderstand something?

 I think I have to read more about reverse mode automatic differentiation…
I think I have to read more about reverse mode automatic differentiation…