Hello!
I’m using NASA’s Ocean color data, chlorophyll to be precise, and a single day of data looks like this:
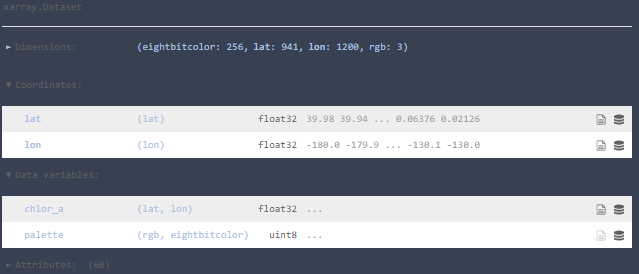
Basically, each pair of (lat,lon) has a single float value of chlor_a. There’s a whole bunch of daily data to be used.
How would I go about feeding this type of .nc data directly into a DCGAN, without reverting to using actual images (.png and .jpeg.)?
Thanks!
You could try to create an “image-like” input tensor in the shape [batch_size, channels, height, width], where the channels could be set to 1, since you are dealing with a single float for each pixel location, and the height and width could be set to the lat and lon.
Thanks! Will give it a try in a few hours, and let you know how it works! Cheers 
[batch_size, channels, height, width]
If I have 100 files of data, each representing a single day values, that would be
[100 (number of daily data/satellite "screenshots"), 1, 941 ( number of lat coordinates), 1200 (number of lon coordinates]
right?
Where do I then input the actual chlor_a values? Sorry if this is a dumb question, I’m very new to all of this and have trouble finding tutorials.
Does that mean that the height, width part is just the x = torch.from_numpy(x) where x = np.array(nc.variables['chlor_a']) and where nc is the “xarray inputed” netCDF file? x already has (lat, lon) dimensions.
In that case I think you can just unsqueeze dim0 and dim1 as x should already contain the values to create the batch and channel dimension.
This would create a tensor in the shape [1, 1, lat, lon] which would be an “image-like” tensor.

