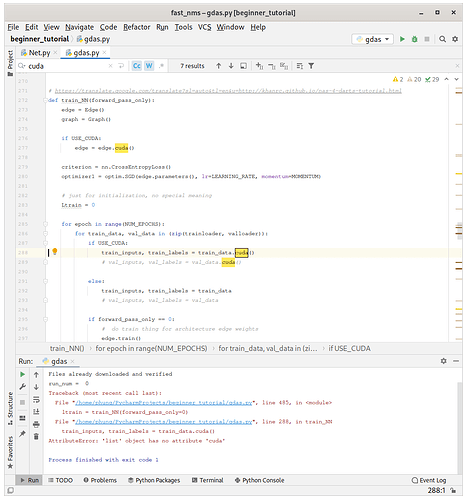
I am having AttributeError: ‘list’ object has no attribute ‘cuda’
To avoid this error, I understand that both train_data and val_data variables should be defined using tensor type.
However, how would I do that exactly without causing other errors ?
# https://github.com/D-X-Y/AutoDL-Projects/issues/99
import torch
import torch.utils.data
import torch.nn as nn
import torch.nn.functional as F
import torch.optim as optim
import torchvision
import torchvision.transforms as transforms
import numpy as np
USE_CUDA = torch.cuda.is_available()
# https://arxiv.org/pdf/1806.09055.pdf#page=12
TEST_DATASET_RATIO = 0.5 # 50 percent of the dataset is dedicated for testing purpose
BATCH_SIZE = 4
NUM_OF_IMAGE_CHANNELS = 3 # RGB
IMAGE_HEIGHT = 32
IMAGE_WIDTH = 32
NUM_OF_IMAGE_CLASSES = 10
SIZE_OF_HIDDEN_LAYERS = 64
NUM_EPOCHS = 50
LEARNING_RATE = 0.025
MOMENTUM = 0.9
NUM_OF_CELLS = 8
NUM_OF_MIXED_OPS = 5
NUM_OF_PREVIOUS_CELLS_OUTPUTS = 2 # last_cell_output , second_last_cell_output
NUM_OF_NODES_IN_EACH_CELL = 4
NUM_OF_CONNECTIONS_PER_CELL = NUM_OF_PREVIOUS_CELLS_OUTPUTS + NUM_OF_NODES_IN_EACH_CELL
NUM_OF_CHANNELS = 16
INTERVAL_BETWEEN_REDUCTION_CELLS = 3
PREVIOUS_PREVIOUS = 2 # (n-2)
REDUCTION_STRIDE = 2
NORMAL_STRIDE = 1
TAU_GUMBEL = 0.5
EDGE_WEIGHTS_NETWORK_IN_SIZE = 5
EDGE_WEIGHTS_NETWORK_OUT_SIZE = 2
# https://pytorch.org/tutorials/beginner/blitz/cifar10_tutorial.html
transform = transforms.Compose(
[transforms.ToTensor(),
transforms.Normalize((0.5, 0.5, 0.5), (0.5, 0.5, 0.5))])
trainset = torchvision.datasets.CIFAR10(root='./data', train=True,
download=True, transform=transform)
trainloader = torch.utils.data.DataLoader(trainset, batch_size=BATCH_SIZE,
shuffle=True, num_workers=2)
valset = torchvision.datasets.CIFAR10(root='./data', train=False,
download=True, transform=transform)
valloader = torch.utils.data.DataLoader(valset, batch_size=BATCH_SIZE,
shuffle=False, num_workers=2)
classes = ('plane', 'car', 'bird', 'cat',
'deer', 'dog', 'frog', 'horse', 'ship', 'truck')
TRAIN_BATCH_SIZE = int(len(trainset) * (1 - TEST_DATASET_RATIO))
# https://discordapp.com/channels/687504710118146232/703298739732873296/853270183649083433
# for training for edge weights as well as internal NN function weights
class Edge(nn.Module):
def __init__(self):
super(Edge, self).__init__()
# https://stackoverflow.com/a/51027227/8776167
# self.linear = nn.Linear(EDGE_WEIGHTS_NETWORK_IN_SIZE, EDGE_WEIGHTS_NETWORK_OUT_SIZE)
# https://pytorch.org/docs/stable/generated/torch.nn.parameter.Parameter.html
self.weights = nn.Parameter(torch.zeros(1),
requires_grad=True) # for edge weights, not for internal NN function weights
def __freeze_w(self):
self.weights.requires_grad = False
def __unfreeze_w(self):
self.weights.requires_grad = True
def __freeze_f(self):
for param in self.f.parameters():
param.requires_grad = False
def __unfreeze_f(self):
for param in self.f.parameters():
param.requires_grad = True
# for NN functions internal weights training
def forward_f(self, x):
self.__unfreeze_f()
self.__freeeze_w()
# inheritance in python classes and SOLID principles
# https://en.wikipedia.org/wiki/SOLID
# https://blog.cleancoder.com/uncle-bob/2020/10/18/Solid-Relevance.html
return self.f(x)
# self-defined initial NAS architecture, for supernet architecture edge weight training
def forward_edge(self, x):
self.__freeze_f()
self.__unfreeze_w()
return x * self.weights
class ConvEdge(Edge):
def __init__(self, stride):
super().__init__()
self.f = nn.Conv2d(in_channels=3, out_channels=6, kernel_size=(3, 3), stride=(stride, stride))
class LinearEdge(Edge):
def __init__(self):
super().__init__()
self.f = nn.Linear(84, 10)
class MaxPoolEdge(Edge):
def __init__(self):
super().__init__()
self.f = nn.MaxPool2d(kernel_size=3)
class AvgPoolEdge(Edge):
def __init__(self):
super().__init__()
self.f = nn.AvgPool2d(kernel_size=3)
class Skip(nn.Module):
def forward(self, x):
return x
class SkipEdge(Edge):
def __init__(self):
super().__init__()
self.f = Skip()
# to collect and manage different edges between 2 nodes
class Connection:
def __init__(self, stride):
super(Connection, self).__init__()
# creates distinct edges and references each of them in a list (self.edges)
self.linear_edge = LinearEdge()
self.conv2d_edge = ConvEdge(stride)
self.maxpool_edge = MaxPoolEdge()
self.avgpool_edge = AvgPoolEdge()
self.skip_edge = SkipEdge()
self.edges = [self.linear_edge, self.conv2d_edge, self.maxpool_edge, self.avgpool_edge, self.skip_edge]
self.edge_weights = torch.zeros(NUM_OF_MIXED_OPS)
# for approximate architecture gradient
self.f_weights = [None] * NUM_OF_MIXED_OPS
self.f_weights_backup = [None] * NUM_OF_MIXED_OPS
self.weight_plus = torch.zeros(NUM_OF_MIXED_OPS)
self.weight_minus = torch.zeros(NUM_OF_MIXED_OPS)
# use linear transformation (weighted summation) to combine results from different edges
self.combined_feature_map = torch.zeros([BATCH_SIZE, NUM_OF_IMAGE_CHANNELS, IMAGE_HEIGHT, IMAGE_WIDTH])
for e in range(NUM_OF_MIXED_OPS):
self.edge_weights[e] = self.edges[e].weights
# https://stackoverflow.com/a/45024500/8776167 extracts the weights learned through NN functions
self.f_weights[e] = list(self.edges[e].parameters())
# Refer to GDAS equations (5) and (6)
# if one_hot is already there, would summation be required given that all other entries are forced to 0 ?
# It's not required, but you don't know, which index is one hot encoded 1.
# https://pytorch.org/docs/stable/nn.functional.html#gumbel-softmax
gumbel = F.gumbel_softmax(self.edge_weights, tau=TAU_GUMBEL, hard=True)
self.chosen_edge = np.argmax(gumbel.detach().numpy(), axis=0) # converts one-hot encoding into integer
# to collect and manege multiple different connections between a particular node and its neighbouring nodes
class Node:
def __init__(self, stride):
super(Node, self).__init__()
# two types of output connections
# Type 1: (multiple edges) output connects to the input of the other intermediate nodes
# Type 2: (single edge) output connects directly to the final output node
# Type 1
self.connections = [Connection(stride) for i in range(NUM_OF_CONNECTIONS_PER_CELL)]
# Type 2
# depends on PREVIOUS node's Type 1 output
self.output = torch.zeros(NUM_OF_IMAGE_CLASSES) # for initialization
# to manage all nodes within a cell
class Cell:
def __init__(self, stride):
super(Cell, self).__init__()
# all the coloured edges inside
# https://user-images.githubusercontent.com/3324659/117573177-20ea9a80-b109-11eb-9418-16e22e684164.png
# A single cell contains 'NUM_OF_NODES_IN_EACH_CELL' distinct nodes
# for the k-th node, we have (k+1) preceding nodes.
# Each intermediate state, 0->3 ('NUM_OF_NODES_IN_EACH_CELL-1'),
# is connected to each previous intermediate state
# as well as the output of the previous two cells, c_{k-2} and c_{k-1} (after a preprocessing layer).
# previous_previous_cell_output = c_{k-2}
# previous_cell_output = c{k-1}
self.nodes = [Node(stride) for i in range(NUM_OF_NODES_IN_EACH_CELL)]
# just for variables initialization
self.previous_cell = 0
self.previous_previous_cell = 0
self.output = torch.zeros(NUM_OF_NODES_IN_EACH_CELL, NUM_OF_IMAGE_CLASSES)
for n in range(NUM_OF_NODES_IN_EACH_CELL):
# 'add' then 'concat' feature maps from different nodes
# needs to take care of tensor dimension mismatch
# See https://github.com/D-X-Y/AutoDL-Projects/issues/99#issuecomment-869100416
self.output += self.nodes[n].output
# to manage all nodes
class Graph:
def __init__(self):
super(Graph, self).__init__()
stride = 0 # just to initialize a variable
for i in range(NUM_OF_CELLS):
if i % INTERVAL_BETWEEN_REDUCTION_CELLS == 0:
stride = REDUCTION_STRIDE # to emulate reduction cell by using normal cell with stride=2
else:
stride = NORMAL_STRIDE # normal cell
self.cells = [Cell(stride) for i in range(NUM_OF_CELLS)]
# https://www.reddit.com/r/learnpython/comments/no7btk/how_to_carry_extra_information_across_dag/
# https://docs.python.org/3/tutorial/datastructures.html
# generates a supernet consisting of 'NUM_OF_CELLS' cells
# each cell contains of 'NUM_OF_NODES_IN_EACH_CELL' nodes
# refer to PNASNet https://arxiv.org/pdf/1712.00559.pdf#page=5 for the cell arrangement
# https://pytorch.org/tutorials/beginner/examples_autograd/two_layer_net_custom_function.html
# encodes the cells and nodes arrangement in the multigraph
for c in range(NUM_OF_CELLS):
if c > 1: # for previous_previous_cell, (c-2)
self.cells[c].previous_cell = self.cells[c-1].output
self.cells[c].previous_previous_cell = self.cells[c - PREVIOUS_PREVIOUS].output
for n in range(NUM_OF_NODES_IN_EACH_CELL):
if n > 0:
# depends on PREVIOUS node's Type 1 connection
# needs to take care tensor dimension mismatch from multiple edges connections
self.cells[c].nodes[n].output = self.cells[c].nodes[n - 1].connections
else: # n == 0
if c > 1: # there is no input from previous cells for the first two cells
# needs to take care tensor dimension mismatch from multiple edges connections
self.cells[c].nodes[n].output = self.cells[c].nodes[n - 1].connections + \
self.cells[c-1].nodes[NUM_OF_NODES_IN_EACH_CELL-1].connections + \
self.cells[c-PREVIOUS_PREVIOUS].nodes[NUM_OF_NODES_IN_EACH_CELL-1].connections
# https://translate.google.com/translate?sl=auto&tl=en&u=http://khanrc.github.io/nas-4-darts-tutorial.html
def train_NN(forward_pass_only):
edge = Edge()
graph = Graph()
if USE_CUDA:
edge = edge.cuda()
criterion = nn.CrossEntropyLoss()
optimizer1 = optim.SGD(edge.parameters(), lr=LEARNING_RATE, momentum=MOMENTUM)
# just for initialization, no special meaning
Ltrain = 0
for epoch in range(NUM_EPOCHS):
for train_data, val_data in (zip(trainloader, valloader)):
if USE_CUDA:
train_inputs, train_labels = train_data.cuda()
# val_inputs, val_labels = val_data.cuda()
else:
train_inputs, train_labels = train_data
# val_inputs, val_labels = val_data
if forward_pass_only == 0:
# do train thing for architecture edge weights
edge.train()
# zero the parameter gradients
optimizer1.zero_grad()
# forward pass
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
for e in range(NUM_OF_MIXED_OPS):
if c == 0:
x = train_inputs
else:
# Uses feature map output from previous neighbour node for further processing
x = graph.cells[c].nodes[n-1].connections[cc].combined_feature_map
# combines all the feature maps from different mixed ops edges
graph.cells[c].nodes[n].connections[cc].combined_feature_map += \
graph.cells[c].nodes[n].connections[cc].edges[e].forward_edge(x) # Ltrain(w±, alpha)
outputs1 = graph.cells[NUM_OF_CELLS-1].output
Ltrain = criterion(outputs1, train_labels)
if forward_pass_only == 0:
# backward pass
Ltrain = Ltrain.requires_grad_()
Ltrain.backward()
optimizer1.step()
else:
# no need to save model parameters for next epoch
return Ltrain
# DARTS's approximate architecture gradient. Refer to equation (8)
# needs to save intermediate trained model for Ltrain
path = './model.pth'
torch.save(graph, path)
return Ltrain
def train_architecture(forward_pass_only, train_or_val='val'):
edge = Edge()
graph = Graph()
if USE_CUDA:
edge = edge.cuda()
criterion = nn.CrossEntropyLoss()
optimizer2 = optim.SGD(edge.parameters(), lr=LEARNING_RATE, momentum=MOMENTUM)
# just for initialization, no special meaning
Lval = 0
for epoch in range(NUM_EPOCHS):
for i, train_data, j, val_data in enumerate(zip(trainloader, valloader)):
if USE_CUDA:
train_inputs, train_labels = train_data.cuda()
val_inputs, val_labels = val_data.cuda()
else:
train_inputs, train_labels = train_data
val_inputs, val_labels = val_data
if forward_pass_only == 0:
# do train thing for internal NN function weights
edge.train()
# zero the parameter gradients
optimizer2.zero_grad()
# forward pass
# use linear transformation ('weighted sum then concat') to combine results from different nodes
# into an output feature map to be fed into the next neighbour node for further processing
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
for e in range(NUM_OF_MIXED_OPS):
x = 0 # depends on the input tensor dimension requirement
if c == 0:
if train_or_val == 'val':
x = val_inputs
else:
x = train_inputs
else:
# Uses feature map output from previous neighbour node for further processing
x = graph.cells[c].nodes[n-1].connections[cc].combined_feature_map
# need to take care of tensors dimension mismatch
graph.cells[c].nodes[n].connections[cc].combined_feature_map += \
graph.cells[c].nodes[n].connections[cc].edge_weights[e] * \
graph.cells[c].nodes[n].connections[cc].edges[e].forward_f(x) # Lval(w*, alpha)
outputs2 = graph.cells[NUM_OF_CELLS-1].output
if train_or_val == 'val':
loss = criterion(outputs2, val_labels)
else:
loss = criterion(outputs2, train_labels)
if forward_pass_only == 0:
# backward pass
Lval = loss
Lval = Lval.requires_grad_()
Lval.backward()
optimizer2.step()
else:
# no need to save model parameters for next epoch
return loss
# DARTS's approximate architecture gradient. Refer to equation (8)
# needs to save intermediate trained model for Lval
path = './model.pth'
torch.save(graph, path)
sigma = LEARNING_RATE
epsilon = 0.01 / torch.norm(Lval)
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
CC = graph.cells[c].nodes[n].connections[cc]
for e in range(NUM_OF_MIXED_OPS):
for w in graph.cells[c].nodes[n].connections[cc].edges[e].parameters():
# https://mythrex.github.io/math_behind_darts/
# Finite Difference Method
CC.weight_plus[e] = w + epsilon * Lval
CC.weight_minus[e] = w - epsilon * Lval
# Backups original f_weights
CC.f_weights_backup[e] = w
# replaces f_weights with weight_plus before NN training
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
CC = graph.cells[c].nodes[n].connections[cc]
for e in range(NUM_OF_MIXED_OPS):
for w in graph.cells[c].nodes[n].connections[cc].edges[e].parameters():
w = CC.weight_plus[e]
# test NN to obtain loss
Ltrain_plus = train_architecture(forward_pass_only=1, train_or_val='train')
# replaces f_weights with weight_minus before NN training
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
CC = graph.cells[c].nodes[n].connections[cc]
for e in range(NUM_OF_MIXED_OPS):
for w in graph.cells[c].nodes[n].connections[cc].edges[e].parameters():
w = CC.weight_minus[e]
# test NN to obtain loss
Ltrain_minus = train_architecture(forward_pass_only=1, train_or_val='train')
# Restores original f_weights
for c in range(NUM_OF_CELLS):
for n in range(NUM_OF_NODES_IN_EACH_CELL):
for cc in range(NUM_OF_CONNECTIONS_PER_CELL):
CC = graph.cells[c].nodes[n].connections[cc]
for e in range(NUM_OF_MIXED_OPS):
for w in graph.cells[c].nodes[n].connections[cc].edges[e].parameters():
w = CC.f_weights_backup[e]
L2train_Lval = (Ltrain_plus - Ltrain_minus) / (2 * epsilon)
return Lval - L2train_Lval
if __name__ == "__main__":
run_num = 0
not_converged = 1
while not_converged:
print("run_num = ", run_num)
ltrain = train_NN(forward_pass_only=0)
lval = train_architecture(forward_pass_only=0, train_or_val='val')
not_converged = (lval > 0.1) or (ltrain > 0.1)
run_num = run_num + 1
# do test thing